6Y4Q
 
 | | Structure of a stapled peptide bound to MDM2 | | Descriptor: | ACE-LEU-THR-PHE-GLY-GLU-TYR-TRP-ALA-GLN-LEU-ALA-SER, E3 ubiquitin-protein ligase Mdm2, ~{N}-[(1-ethyl-1,2,3-triazol-4-yl)methyl]-~{N},5-dimethyl-4-[2-[2-methyl-5-[methyl-[(1-propyl-1,2,3-triazol-4-yl)methyl]carbamoyl]thiophen-3-yl]cyclopenten-1-yl]thiophene-2-carboxamide | | Authors: | Pantelejevs, T, Bakanovych, I. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Diarylethene moiety as an enthalpy-entropy switch: photoisomerizable stapled peptides for modulating p53/MDM2 interaction.
Org.Biomol.Chem., 18, 2020
|
|
6TUU
 
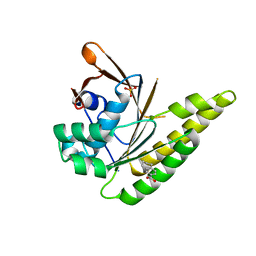 | | Leishmania infantum Rad51 surrogate LiRadA10 in complex with 5,6,7,8-tetrahydro-2-naphthoic acid | | Descriptor: | 5,6,7,8-tetrahydronaphthalene-2-carboxylic acid, CHLORIDE ION, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of dedicated crystallographic systems for structure-guided drug discovery
To Be Published
|
|
7NUF
 
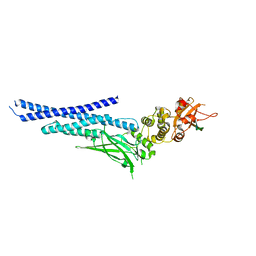 | | Vaccinia virus protein 018 in complex with STAT1 | | Descriptor: | ACETYL GROUP, SULFATE ION, Signal transducer and activator of transcription 1-alpha/beta, ... | | Authors: | Pantelejevs, T, Talbot-Cooper, C, Smith, G.L, Hyvonen, M. | | Deposit date: | 2021-03-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.0004015 Å) | | Cite: | Poxviruses and paramyxoviruses use a conserved mechanism of STAT1 antagonism to inhibit interferon signaling.
Cell Host Microbe, 30, 2022
|
|
3M8Y
 
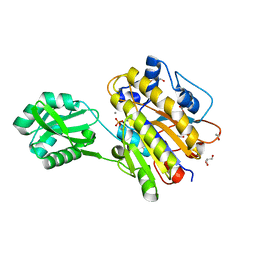 | | Phosphopentomutase from Bacillus cereus after glucose-1,6-bisphosphate activation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3M8Z
 
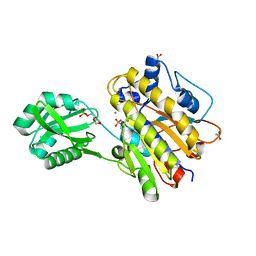 | | Phosphopentomutase from Bacillus cereus bound with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3M8W
 
 | | Phosphopentomutase from Bacillus cereus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
8BR9
 
 | |
8C3N
 
 | |
7QV8
 
 | |
6HQU
 
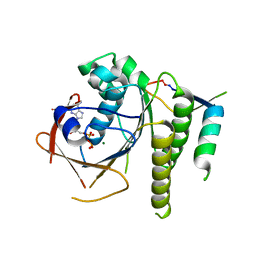 | | Humanised RadA mutant HumRadA22 in complex with a recombined BRC repeat 8-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Breast cancer type 2 susceptibility, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Lindenburg, L, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Improved RAD51 binders through motif shuffling based on the modularity of BRC repeats.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8C3J
 
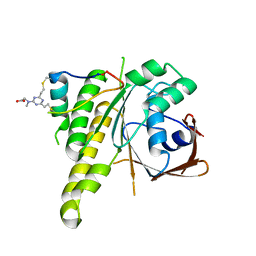 | | Stapled peptide SP2 in complex with humanised RadA mutant HumRadA22 | | Descriptor: | 2-[(4,6-diethyl-1,3,5-triazin-2-yl)-methyl-amino]ethanoic acid, Breast cancer type 2 susceptibility protein, DNA repair and recombination protein RadA | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2022-12-26 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | A recombinant approach for stapled peptide discovery yields inhibitors of the RAD51 recombinase.
Chem Sci, 14, 2023
|
|
5V47
 
 | | Crystal structure of the SR1 domain of lizard sacsin | | Descriptor: | Lizard sacsin, SULFATE ION | | Authors: | Pan, T, Menade, M, Kozlov, G, Gehring, K. | | Deposit date: | 2017-03-08 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of ubiquitin-like (Ubl) and Hsp90-like domains of sacsin provide insight into pathological mutations.
J. Biol. Chem., 293, 2018
|
|
3OT9
 
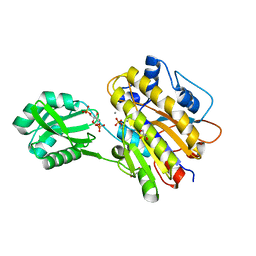 | | Phosphopentomutase from Bacillus cereus bound to glucose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Phalen, V, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-09-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
3TWZ
 
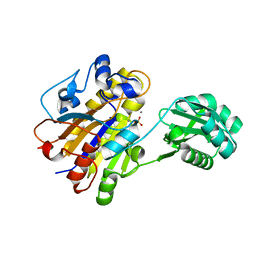 | | Phosphorylated Bacillus cereus phosphopentomutase in space group P212121 | | Descriptor: | MANGANESE (II) ION, Phosphopentomutase | | Authors: | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-02-29 | | Last modified: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3TX0
 
 | | Unphosphorylated Bacillus cereus phosphopentomutase in a P212121 crystal form | | Descriptor: | MANGANESE (II) ION, Phosphopentomutase | | Authors: | Panosian, T.P, Nanneman, D.P, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2011-09-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
3Q1R
 
 | | Crystal structure of a bacterial RNase P holoenzyme in complex with TRNA and in the presence of 5' leader | | Descriptor: | MAGNESIUM ION, RNase P RNA, Ribonuclease P protein component, ... | | Authors: | Reiter, N.J, Ostermanm, A, Torres-Larios, A, Swinger, K.K, Pan, T, Mondragon, A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.21 Å) | | Cite: | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA.
Nature, 468, 2010
|
|
3Q1Q
 
 | | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, RNase P RNA, ... | | Authors: | Reiter, N.J, Osterman, A, Torres-Larios, A, Swinger, K.K, Pan, T, Mondragon, A. | | Deposit date: | 2010-12-17 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA.
Nature, 468, 2010
|
|
2A2E
 
 | | Crystal structure of the RNA subunit of Ribonuclease P. Bacterial A-type. | | Descriptor: | OSMIUM ION, RNA subunit of RNase P | | Authors: | Torres-Larios, A, Swinger, K.K, Krasilnikov, A.S, Pan, T, Mondragon, A. | | Deposit date: | 2005-06-22 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Crystal structure of the RNA component of bacterial ribonuclease P.
Nature, 437, 2005
|
|
1U9S
 
 | | Crystal structure of the specificity domain of Ribonuclease P of the A-type | | Descriptor: | BARIUM ION, RIBONUCLEASE P | | Authors: | Krasilnikov, A.S, Xiao, Y, Pan, T, Mondragon, A. | | Deposit date: | 2004-08-10 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Basis for structural diversity in homologous RNAs.
Science, 306, 2004
|
|
1NBS
 
 | | Crystal structure of the specificity domain of Ribonuclease P RNA | | Descriptor: | LEAD (II) ION, MAGNESIUM ION, RIBONUCLEASE P RNA | | Authors: | Krasilnikov, A.S, Yang, X, Pan, T, Mondragon, A. | | Deposit date: | 2002-12-03 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of the specificity domain of Ribonuclease P
Nature, 421, 2003
|
|
6B3Y
 
 | | Crystal structure of the PH-like domain from DENND3 | | Descriptor: | DENN domain-containing protein 3 | | Authors: | Kozlov, G, Xu, J, Menade, M, Beaugrand, M, Pan, T, McPherson, P.S, Gehring, K. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | A PH-like domain of the Rab12 guanine nucleotide exchange factor DENND3 binds actin and is required for autophagy.
J. Biol. Chem., 293, 2018
|
|
3GB7
 
 | | Potassium Channel KcsA-Fab complex in Li+ | | Descriptor: | DIACYL GLYCEROL, NICKEL (II) ION, Voltage-gated potassium channel, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-02-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3IGA
 
 | | Potassium Channel KcsA-Fab complex in Li+ and K+ | | Descriptor: | Antibody Fab Fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Thompson, A.N, Ilsoo, K, Panosian, T.D, Iverson, T.M, Allen, T.W, Nimigean, C.M. | | Deposit date: | 2009-07-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mechanism of potassium-channel selectivity revealed by Na(+) and Li(+) binding sites within the KcsA pore.
Nat.Struct.Mol.Biol., 16, 2009
|
|
7ZWD
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U5 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZX7
 
 | | Structure of SNAPc containing Pol II pre-initiation complex bound to U1 snRNA promoter (CC) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Rengachari, S, Schilbach, S, Kaliyappan, T, Gouge, J, Zumer, K, Schwarz, J, Urlaub, H, Dienemann, C, Vannini, A, Cramer, P. | | Deposit date: | 2022-05-20 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Nat.Struct.Mol.Biol., 29, 2022
|
|
