3TL4
 
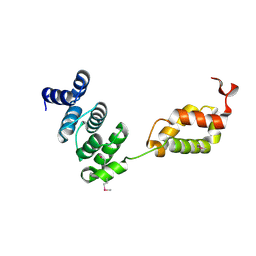 | |
3J9S
 
 | |
2VY9
 
 | | Molecular architecture of the stressosome, a signal integration and transduction hub | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST | | Authors: | Marles-Wright, J, Grant, T, Delumeau, O, van Duinen, G, Firbank, S.J, Lewis, P.J, Murray, J.W, Newman, J.A, Quin, M.B, Race, P.R, Rohou, A, Tichelaar, W, van Heel, M, Lewis, R.J. | | Deposit date: | 2008-07-21 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular Architecture of the "Stressosome," a Signal Integration and Transduction Hub
Science, 322, 2008
|
|
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FMB
 
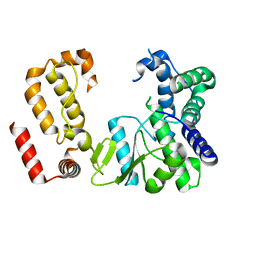 | | Nodavirus RNA replication protein A polymerase domain, local refinement | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FAK
 
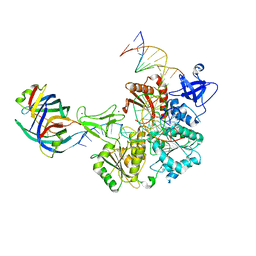 | | DNA replication fork binding triggers structural changes in the PriA DNA helicase that regulate the PriA-PriB replication restart pathway in E. coli | | Descriptor: | DNA (5'-D(P*CP*AP*GP*AP*CP*TP*CP*AP*TP*TP*TP*AP*GP*CP*CP*CP*TP*TP*AP*TP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*AP*TP*AP*AP*GP*GP*GP*CP*TP*GP*AP*GP*CP*AP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*GP*CP*GP*TP*GP*CP*TP*C)-3'), ... | | Authors: | Duckworth, A.T, Ducos, P.L, McMillan, S.D, Satyshur, K.A, Blumenthal, K.H, Deorio, H.R, Larson, J.A, Sandler, S.J, Grant, T, Keck, J.L. | | Deposit date: | 2022-11-28 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Replication fork binding triggers structural changes in the PriA helicase that govern DNA replication restart in E. coli.
Nat Commun, 14, 2023
|
|
8FWA
 
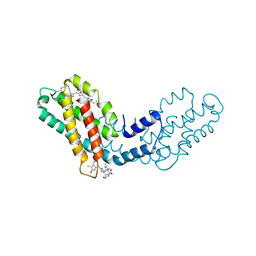 | | Phycocyanin structure from a modular droplet injector for serial femtosecond crystallography | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN, ... | | Authors: | Botha, S, Doppler, D.L, Sonker, M, Egatz-Gomez, A, Grieco, A, Zaare, S, Jernigan, R, Meza-Aguilar, J.D, Rabbani, M.T, Manna, A, Alvarez, R, Karpos, K, Cruz Villarreal, J, Nelson, G, Ketawala, G.K, Pey, A.L, Ruiz-Fresneda, M.A, Pacheco-Garcia, J.L, Nazari, R, Sierra, R, Hunter, M.S, Batyuk, A, Kupitz, C.J, Sublett, R.E, Lisova, S, Mariani, V, Boutet, S, Fromme, R, Grant, T.D, Fromme, P, Kirian, R.A, Martin-Garcia, J.M, Ros, A. | | Deposit date: | 2023-01-20 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Modular droplet injector for sample conservation providing new structural insight for the conformational heterogeneity in the disease-associated NQO1 enzyme.
Lab Chip, 23, 2023
|
|
6PNY
 
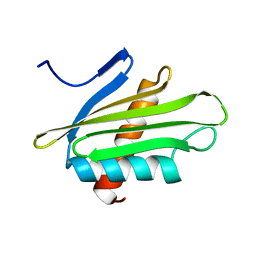 | | X-ray Structure of Flpp3 | | Descriptor: | Flpp3 | | Authors: | Zook, J.D, Shekhar, M, Hansen, D.T, Conrad, C, Grant, T.D, Gupta, C, White, T, Barty, A, Basu, S, Zhao, Y, Zatsepin, N.A, Ishchenko, A, Batyuk, A, Gati, C, Li, C, Galli, L, Coe, J, Hunter, M, Liang, M, Weierstall, U, Nelson, G, James, D, Stauch, B, Craciunescu, F, Thifault, D, Liu, W, Cherezov, V, Singharoy, A, Fromme, P. | | Deposit date: | 2019-07-03 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | XFEL and NMR Structures of Francisella Lipoprotein Reveal Conformational Space of Drug Target against Tularemia.
Structure, 28, 2020
|
|
8FWJ
 
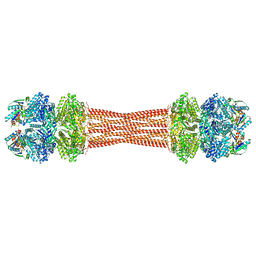 | | Structure of dodecameric KaiC-RS-S413E/S414E complexed with KaiB-RS solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiB, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6OJ6
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase, Template, ... | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
8FWI
 
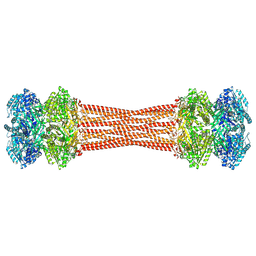 | | Structure of dodecameric KaiC-RS-S413E/S414E solved by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein KaiC, ... | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2023-01-22 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
2M89
 
 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
4H3S
 
 | | The Structure of Glutaminyl-tRNA Synthetase from Saccharomyces Cerevisiae | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, BROMIDE ION, ... | | Authors: | Snell, E.H, Grant, T.D. | | Deposit date: | 2012-09-14 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Yeast Glutaminyl-tRNA Synthetase and Modeling of Its Interaction with tRNA.
J.Mol.Biol., 425, 2013
|
|
8DBA
 
 | | Crystal structure of dodecameric KaiC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circadian clock protein KaiC, MAGNESIUM ION | | Authors: | Padua, R.A.P, Grant, T, Pitsawong, W, Hoemberger, M.S, Otten, R, Bradshaw, N, Grigorieff, N, Kern, D. | | Deposit date: | 2022-06-14 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | From primordial clocks to circadian oscillators.
Nature, 616, 2023
|
|
6OJ3
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6OUT
 
 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4CRZ
 
 | | Direct visualisation of strain-induced protein prost-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4CS0
 
 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the Pand/Panz Protein Complex Reveals Negative Feedback Regulation of Pantothenate Biosynthesis by Coenzyme A.
Chem.Biol., 22, 2015
|
|
4CRY
 
 | | Direct visualisation of strain-induced protein post-translational modification | | Descriptor: | ACETYL COENZYME *A, ASPARTATE 1-DECARBOXYLASE, CHLORIDE ION, ... | | Authors: | Monteiro, D.C.F, Patel, V, Bartlett, C.P, Grant, T.D, Nozaki, S, Gowdy, J.A, Snell, E.H, Niki, H, Pearson, A.R, Webb, M.E. | | Deposit date: | 2014-03-02 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Direct Visualisation of Strain-Induced Protein Post-Translational Modification
To be Published
|
|
6N1Q
 
 | | Dihedral oligomeric complex of GyrA N-terminal fragment, solved by cryoEM in D2 symmetry | | Descriptor: | DNA gyrase subunit A | | Authors: | Soczek, K.M, Grant, T, Rosenthal, P.B, Mondragon, A. | | Deposit date: | 2018-11-10 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | CryoEM structures of open dimers of Gyrase A in complex with DNA illuminate mechanism of strand passage.
Elife, 7, 2018
|
|
6OU9
 
 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUC
 
 | | Asymmetric focsued reconstruction of human norovirus GII.2 Snow Mountain Virus strain VLP asymmetric unit in T=1 symmetry | | Descriptor: | Viral protein 1, ZINC ION | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
