6RPG
 
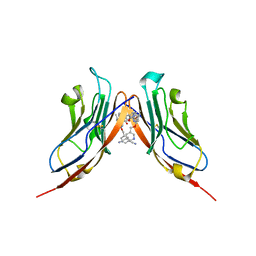 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
3X21
 
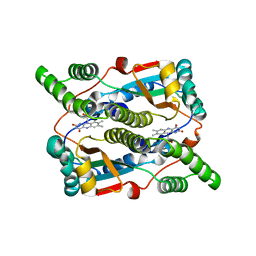 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant T41L/N71S/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Chembiochem, 16, 2015
|
|
3X22
 
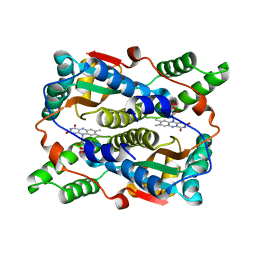 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
8PKP
 
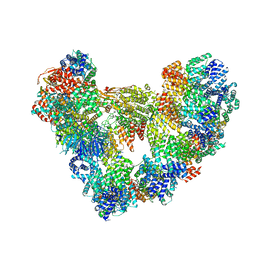 | | Cryo-EM structure of the apo Anaphase-promoting complex/cyclosome (APC/C) at 3.2 Angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Hoefler, A, Yu, J, Chang, L, Zhang, Z, Yang, J, Boland, A, Barford, D. | | Deposit date: | 2023-06-27 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution structure of the Anaphase-promoting complex (APC/C) bound to co-activator Cdh1
To Be Published
|
|
5LCW
 
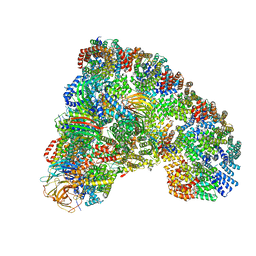 | | Cryo-EM structure of the Anaphase-promoting complex/Cyclosome, in complex with the Mitotic checkpoint complex (APC/C-MCC) at 4.2 angstrom resolution | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Alfieri, C, Chang, L, Zhang, Z, Yang, J, Maslen, S, Skehel, M, Barford, D. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis of APC/C regulation by the spindle assembly checkpoint.
Nature, 536, 2016
|
|
5MZ6
 
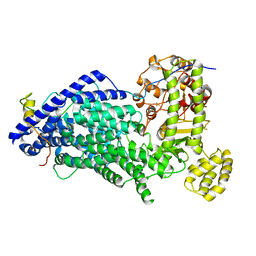 | | Cryo-EM structure of a Separase-Securin complex from Caenorhabditis elegans at 3.8 A resolution | | Descriptor: | Interactor of FizzY protein, SEParase | | Authors: | Boland, A, Martin, T.G, Zhang, Z, Yang, J, Bai, X.C, Chang, L, Scheres, S.H.W, Barford, D. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6J0B
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath-tube complex in extended state | | Descriptor: | Pvc1, Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0C
 
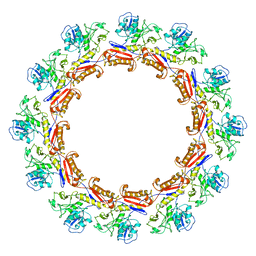 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath complex in contracted state | | Descriptor: | Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0F
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath/tube terminator in extended state | | Descriptor: | Pvc1, Pvc16 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0N
 
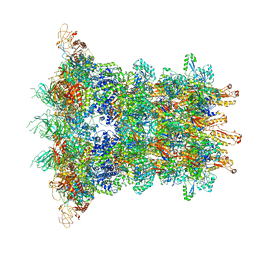 | | Cryo-EM Structure of an Extracellular Contractile Injection System, baseplate in extended state, refined in C6 symmetry | | Descriptor: | Pvc1, Pvc11, Pvc12, ... | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0M
 
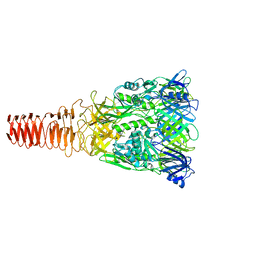 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC baseplate in extended state (reconstructed with C3 symmetry) | | Descriptor: | Pvc8 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
8HNS
 
 | | Crystal structure of an anti-CRISPR protein AcrIIC4 in apo form | | Descriptor: | GLYCEROL, anti-CRISPR protein AcrIIC4 | | Authors: | Sun, W, Cheng, Z, Yang, J, Wang, Y. | | Deposit date: | 2022-12-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | AcrIIC4 inhibits type II-C Cas9 by preventing R-loop formation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4AYT
 
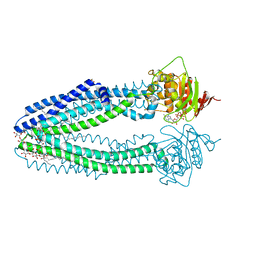 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10 MITOCHONDRIAL, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYX
 
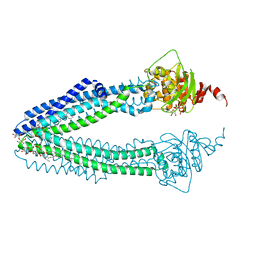 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6JAU
 
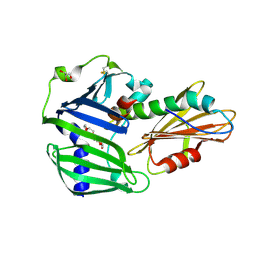 | | The complex structure of Pseudomonas aeruginosa MucA/MucB. | | Descriptor: | CALCIUM ION, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Li, T, He, L.H, Li, C.C, Liu, L, Peng, C.T, Shen, Y.L, Qin, X.F, Xiao, Q.J, Zhu, Y.B, Song, Y.J, Zhao, N.l, Zhao, C, Yang, J, Mu, X.Y, Huang, Q, Bao, R. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Commun Biol, 3, 2020
|
|
6JCJ
 
 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
1NHZ
 
 | | Crystal Structure of the Antagonist Form of Glucocorticoid Receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLUCOCORTICOID RECEPTOR, HEXANE-1,6-DIOL | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
3ZN3
 
 | | N-terminal domain of S. pombe Cdc23 APC subunit | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8, MERCURY (II) ION | | Authors: | Zhang, Z, Yang, J, Conin, N, Kulkarni, K, Barford, D. | | Deposit date: | 2013-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Four Canonical Tpr Subunits of Human Apc/C Form Related Homo-Dimeric Structures and Stack in Parallel to Form a Tpr Suprahelix
J.Mol.Biol., 425, 2013
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
