4XPA
 
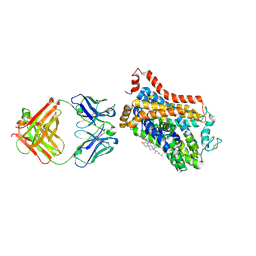 | | X-ray structure of Drosophila dopamine transporter bound to 3,4dichlorophenethylamine | | Descriptor: | 2-(3,4-dichlorophenyl)ethanamine, Antibody fragment heavy chain-protein, 9D5-heavy chain, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1RWT
 
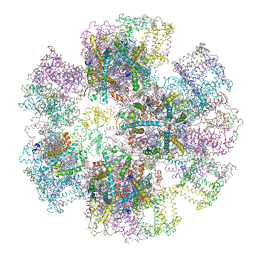 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
6VYI
 
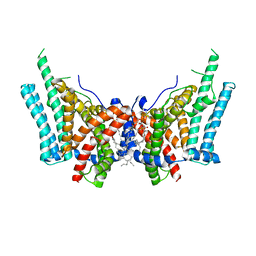 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese, V.R. | | Deposit date: | 2020-02-26 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
6VZ1
 
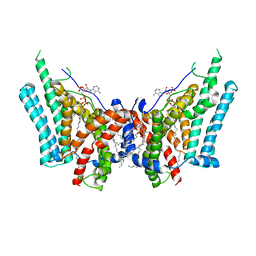 | | Cryo-EM structure of human diacylglycerol O-acyltransferase 1 complexed with acyl-CoA substrate | | Descriptor: | Diacylglycerol O-acyltransferase 1, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Sui, X, Wang, K, Gluchowski, N, Liao, M, Walther, C.T, Farese Jr, V.R. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme.
Nature, 581, 2020
|
|
7C9Z
 
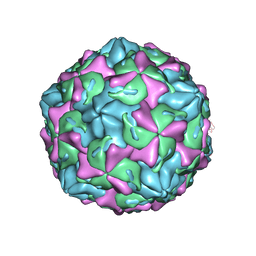 | | Coxsackievirus B1 F-particle | | Descriptor: | MYRISTIC ACID, PALMITIC ACID, VP1, ... | | Authors: | Feng, R, Wang, K, Rao, Z, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
6F7H
 
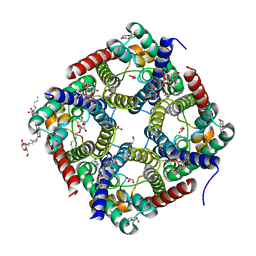 | | Crystal structure of human AQP10 | | Descriptor: | Aquaporin-10, GLYCEROL, nonyl beta-D-glucopyranoside | | Authors: | Gotfryd, K, Wang, K, Missel, J.W, Pedersen, P.A, Gourdon, P. | | Deposit date: | 2017-12-08 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Human adipose glycerol flux is regulated by a pH gate in AQP10.
Nat Commun, 9, 2018
|
|
7F3Q
 
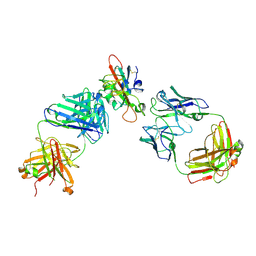 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
1HQW
 
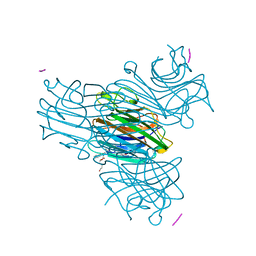 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CONCANAVALIN A WITH A TRIPEPTIDE YPY | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Zhang, Z, Qian, M, Huang, Q, Jia, Y, Tang, Y, Wang, K, Cui, D, Li, M. | | Deposit date: | 2000-12-20 | | Release date: | 2003-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the complex of concanavalin A and tripeptide
J.PROTEIN CHEM., 20, 2001
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
6VH7
 
 | | Doublet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
6USN
 
 | | Co-crystal structure of SPR with compound 5 | | Descriptor: | (2-hydroxyphenyl)[3-methyl-1-(pyridin-2-yl)-1H-pyrazolo[3,4-b]pyridin-5-yl]methanone, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Huang, X, Wang, K. | | Deposit date: | 2019-10-28 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Virtual screening to identify potent sepiapterin reductase inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VHA
 
 | | Singlet Tau Fibril from Corticobasal Degeneration Human Brain Tissue | | Descriptor: | Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-09 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
6VI3
 
 | | Straight Filament from Alzheimer's Disease Human Brain Tissue | | Descriptor: | GLYCINE, Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains
Cell, 180, 2020
|
|
6VHL
 
 | | Paired Helical Filament from Alzheimer's Disease Human Brain Tissue | | Descriptor: | GLYCINE, Microtubule-associated protein tau | | Authors: | Arakhamia, T, Lee, C.E, Carlomagno, Y, Duong, D.M, Kundinger, S.R, Wang, K, Williams, D, DeTure, M, Dickson, D.W, Cook, C.N, Seyfried, N.T, Petrucelli, L, Fitzpatrick, A.W.P. | | Deposit date: | 2020-01-10 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains.
Cell, 180, 2020
|
|
8HAY
 
 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
8WCN
 
 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
