4USJ
 
 | | N-acetylglutamate kinase from Arabidopsis thaliana in complex with PII from Chlamydomonas reinhardtii | | Descriptor: | ACETYLGLUTAMATE KINASE, CHLOROPLASTIC, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
8ARM
 
 | | Crystal structure of LSSmScarlet2 | | Descriptor: | LSSmScarlet2, SULFATE ION | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Subach, F.V. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | LSSmScarlet2 and LSSmScarlet3, Chemically Stable Genetically Encoded Red Fluorescent Proteins with a Large Stokes' Shift.
Int J Mol Sci, 23, 2022
|
|
1WBF
 
 | | WINGED BEAN LECTIN, SACCHARIDE FREE FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Manoj, N, Srinivas, V.R, Suguna, K. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of basic winged-bean lectin and a comparison with its saccharide-bound form.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5I84
 
 | |
1JUU
 
 | | NMR Structure of a Parallel Stranded DNA Duplex at Atomic Resolution | | Descriptor: | 5'-D(P*CP*CP*AP*TP*AP*AP*TP*TP*TP*AP*CP*C)-3', 5'-D(P*CP*CP*TP*AP*TP*TP*AP*AP*AP*TP*CP*C)-3' | | Authors: | Parvathy, V.R, Bhaumik, S.R, Chary, K.V.R, Govil, G, Liu, K, Howard, F.B, Miles, H.T. | | Deposit date: | 2001-08-28 | | Release date: | 2002-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a parallel-stranded DNA duplex at atomic resolution.
Nucleic Acids Res., 30, 2002
|
|
4USI
 
 | | Nitrogen regulatory protein PII from Chlamydomonas reinhardtii in complex with MgATP and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chellamuthu, V.R, Forchhammer, K, Hartmann, M.D. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A Widespread Glutamine-Sensing Mechanism in the Plant Kingdom.
Cell(Cambridge,Mass.), 159, 2014
|
|
5UUY
 
 | | Crystal structure of Dioclea lasiocarpa lectin (DLL) complexed with X-MAN | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Dioclea lasiocarpa lectin (DLL), ... | | Authors: | Santiago, M.Q, Pinto-Junior, V.R, Osterne, V.J.S, Rocha, C.R.C, Neco, A.H.B, Fonseca, F.M.P, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2017-02-17 | | Release date: | 2017-10-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural analysis of Dioclea lasiocarpa lectin: A C6 cells apoptosis-inducing protein.
Int. J. Biochem. Cell Biol., 92, 2017
|
|
4USH
 
 | |
5KDI
 
 | | How FAPP2 Selects Simple Glycosphingolipids Using the GLTP-fold | | Descriptor: | (~{Z})-~{N}-[(~{E},2~{S},3~{R})-1-[(2~{R},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-octadec-4-en-2-yl]octadec-9-enamide, Pleckstrin homology domain-containing family A member 8 | | Authors: | Ochoa-Lizarralde, B, Popov, A.N, Samygina, V.R, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2016-06-08 | | Release date: | 2017-12-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family.
J.Biol.Chem., 293, 2018
|
|
3JTM
 
 | | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana | | Descriptor: | AZIDE ION, Formate dehydrogenase, mitochondrial, ... | | Authors: | Timofeev, V.I, Shabalin, I.G, Serov, A.E, Polyakov, K.M, Popov, V.O, Tishkov, V.I, Kuranova, I.P, Samigina, V.R. | | Deposit date: | 2009-09-13 | | Release date: | 2010-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of recombinant formate dehydrogenase from Arabidopsis thaliana
to be published
|
|
8P5F
 
 | | Human wild-type GAPDH,orthorhombic form | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Samygina, V.R, Muronetz, V.I, Schmalhausen, E.V. | | Deposit date: | 2023-05-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | S-nitrosylation and S-glutathionylation of GAPDH: Similarities, differences, and relationships.
Biochim Biophys Acta Gen Subj, 1867, 2023
|
|
2VMX
 
 | | Crystal structure of F351GbsSHMT in complex with L-allo-Thr | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALLO-THREONINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Pai, V.R, Bisht, S, Bhavani, B.S, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-01-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and Functional Studies of Bacillus Stearothermophilus Serine Hydroxymethyltransferase: The Role of Asn(341), Tyr(60) and Phe(351) in Tetrahydrofolate Binding.
Biochem.J., 418, 2009
|
|
2VMP
 
 | | Crystal structure of N341AbsSHMT L-Ser external aldimine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Pai, V.R, Bisht, S, Bhavani, B.S, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-01-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Functional Studies of Bacillus Stearothermophilus Serine Hydroxymethyltransferase: The Role of Asn(341), Tyr(60) and Phe(351) in Tetrahydrofolate Binding.
Biochem.J., 418, 2009
|
|
2VMY
 
 | | Crystal structure of F351GbsSHMT in complex with Gly and FTHF | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, GLYCINE, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Rajaram, V, Pai, V.R, Bisht, S, Bhavani, B.S, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2008-01-29 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Studies of Bacillus Stearothermophilus Serine Hydroxymethyltransferase: The Role of Asn(341), Tyr(60) and Phe(351) in Tetrahydrofolate Binding.
Biochem.J., 418, 2009
|
|
6FVE
 
 | |
6FVH
 
 | |
1JO0
 
 | | Structure of HI1333, a Hypothetical Protein from Haemophilus influenzae with Structural Similarity to RNA-binding Proteins | | Descriptor: | GLYCEROL, HYPOTHETICAL PROTEIN HI1333 | | Authors: | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2001-07-26 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae
Proteins, 49, 2002
|
|
6Z9Z
 
 | | Atomic resolution X-ray structure of the Uridine phosphorylase from Vibrio cholerae on crystals grown under microgravity | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Eistrikh-Heller, P.A, Rubinsky, S.V, Samygina, V.R, Gabdulkhakov, A.G, Lashkov, A.A. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Crystallization in Microgravity and the Atomic-Resolution Structure of Uridine Phosphorylase from Vibrio cholerae
Crystallography Reports, 66, 2021
|
|
5F5Q
 
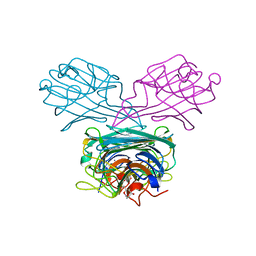 | | Crystal structure of Canavalia virosa lectin in complex with alpha-methyl-mannoside | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Osterne, V.J.S, Silva-Filho, J.C, Pinto-Junior, V.R, Santiago, M.Q, Lossio, C.F, Delatorre, P, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural characterization of a lectin from Canavalia virosa seeds with inflammatory and cytotoxic activities.
Int.J.Biol.Macromol., 94, 2016
|
|
5EYX
 
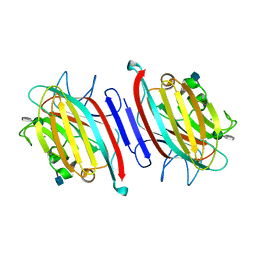 | | Monoclinic Form of Centrolobium tomentosum seed lectin (CTL) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Centrolobium tomentosum lectin, ... | | Authors: | Almeida, A.C, Osterne, V.J.S, Santiago, M.Q, Pinto-Junior, V.R, Silva-Filho, J.C, Lossio, C.F, Almeida, R.P.H, Teixeira, C.S, Delatorre, P, Rocha, B.A.M, Santiago, K.S, Cavada, B.S. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity.
Arch.Biochem.Biophys., 596, 2016
|
|
5EYY
 
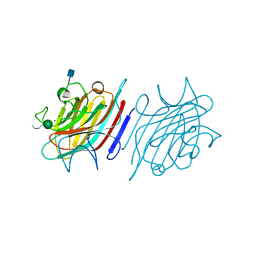 | | Tetragonal Form of Centrolobium tomentosum seed lectin (CTL) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Centrolobium tomentosum lectin, ... | | Authors: | Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Almeida, A.C, Lossio, C.F, Silva-Filho, J.C, Almeida, R.P.H, Teixeira, C.S, Delatorre, P, Rocha, B.A.M, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity.
Arch.Biochem.Biophys., 596, 2016
|
|
2XKJ
 
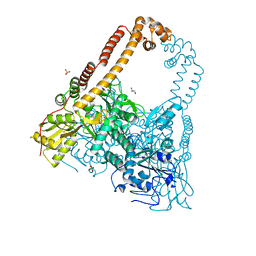 | | CRYSTAL STRUCTURE OF CATALYTIC CORE OF A. BAUMANNII TOPO IV (PARE- PARC FUSION TRUNCATE) | | Descriptor: | GLYCEROL, SULFATE ION, TOPOISOMERASE IV | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XBP
 
 | | A novel signal transduction protein PII variant from Synechococcus elongatus PCC7942 indicates a two-step process for NAGK PII complex formation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fokina, O, Chellamuthu, V.R, Zeth, K, Forchhammer, K. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Novel Signal Transduction Protein P(II) Variant from Synechococcus Elongatus Pcc 7942 Indicates a Two-Step Process for Nagk- P(II) Complex Formation.
J.Mol.Biol., 399, 2010
|
|
2XKK
 
 | | CRYSTAL STRUCTURE OF MOXIFLOXACIN, DNA, and A. BAUMANNII TOPO IV (PARE-PARC FUSION TRUNCATE) | | Descriptor: | 1-cyclopropyl-6-fluoro-8-methoxy-7-[(4aS,7aS)-octahydro-6H-pyrrolo[3,4-b]pyridin-6-yl]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, DNA, MAGNESIUM ION, ... | | Authors: | Wohlkonig, A, Chan, P.F, Fosberry, A.P, Homes, P, Huang, J, Kranz, M, Leydon, V.R, Miles, T.J, Pearson, N.D, Perera, R.L, Shillings, A.J, Gwynn, M.N, Bax, B.D. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis of Quinolone Inhibition of Type Iia Topoisomerases and Target-Mediated Resistance
Nat.Struct.Mol.Biol., 17, 2010
|
|
8FH3
 
 | | Human IFT-A complex structures provide molecular insights into ciliary transport | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 140 homolog, Tubby-related protein 3, ... | | Authors: | Jiang, M, Palicharla, V.R, Miller, D, Hwang, S.H, Zhu, H, Hixson, P, Mukhopadhyay, S, Sun, J. | | Deposit date: | 2022-12-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Human IFT-A complex structures provide molecular insights into ciliary transport.
Cell Res., 33, 2023
|
|
