6V3K
 
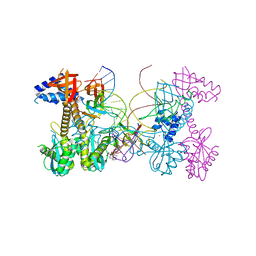 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ419 (compound 4c) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-(5-oxidanylpentyl)-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-11-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
4C3G
 
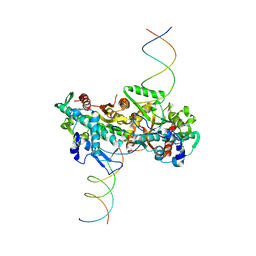 | | cryo-EM structure of activated and oligomeric restriction endonuclease SgrAI | | Descriptor: | 5'-D(*CP*CP*GP*GP*TP*GP*TP*GP*AP*AP*GP*AP*CP*CP *CP*AP*CP*GP*CP*AP*TP*CP)-3', 5'-D(*GP*AP*TP*GP*CP*GP*TP*GP*GP*GP*TP*CP*TP*TP *CP*AP*CP*AP)-3', SGRAIR RESTRICTION ENZYME | | Authors: | Lyumkis, D, Talley, H, Stewart, A, Shah, S, Park, C.K, Tama, F, Potter, C.S, Carragher, B, Horton, N.C. | | Deposit date: | 2013-08-23 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Allosteric Regulation of DNA Cleavage and Sequence-Specificity Through Run-on Oligomerization.
Structure, 21, 2013
|
|
6PUW
 
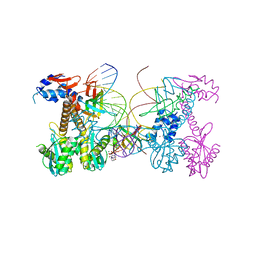 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | Descriptor: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUZ
 
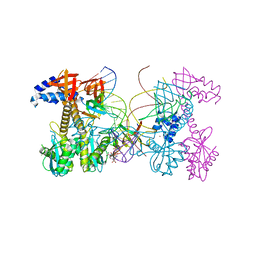 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
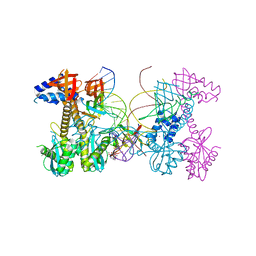 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
7SEP
 
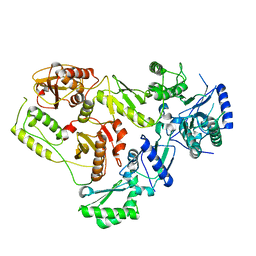 | |
6PUT
 
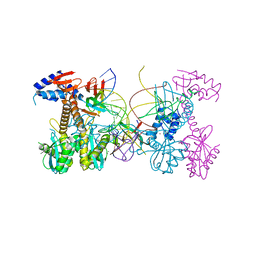 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | | Descriptor: | CALCIUM ION, Chimeric Sso7d and HIV-1 integrase, ZINC ION, ... | | Authors: | Lyumkis, D, Jozwik, I.K, Passos, D. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
7SJX
 
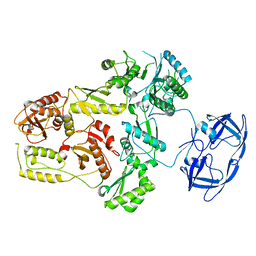 | | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | | Descriptor: | Gag-Pol polyprotein | | Authors: | Lyumkis, D, Passos, D, Arnold, E, Harrison, J.J.E.K, Ruiz, F.X. | | Deposit date: | 2021-10-19 | | Release date: | 2022-07-27 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation.
Sci Adv, 8, 2022
|
|
3J5M
 
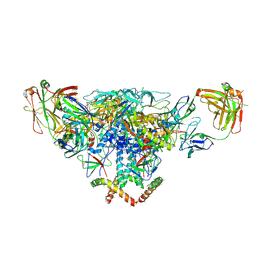 | | Cryo-EM structure of the BG505 SOSIP.664 HIV-1 Env trimer with 3 PGV04 Fabs | | Descriptor: | BG505 SOSIP gp120, BG505 SOSIP gp41, PGV04 heavy chain, ... | | Authors: | Lyumkis, D, Julien, J.-P, Wilson, I.A, Ward, A.B. | | Deposit date: | 2013-10-26 | | Release date: | 2013-11-13 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
5TQW
 
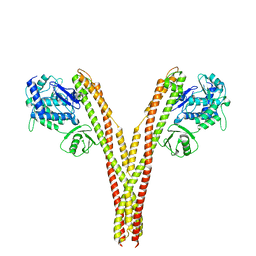 | | CryoEM reconstruction of human IKK1, open conformation 1 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5TQY
 
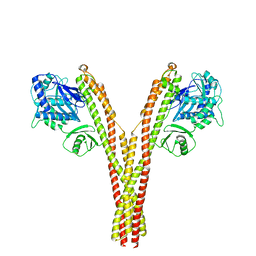 | | CryoEM reconstruction of human IKK1, closed conformation 3 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
5U1C
 
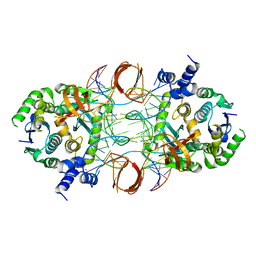 | |
5TQX
 
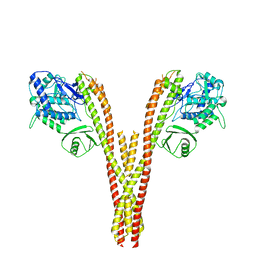 | | CryoEM reconstruction of human IKK1, intermediate conformation 2 | | Descriptor: | Inhibitor of nuclear factor kappa-B kinase subunit alpha | | Authors: | Lyumkis, D, Ghosh, G, Polley, S, Biswath, T, Huang, D, Passos, D.O. | | Deposit date: | 2016-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
6ME0
 
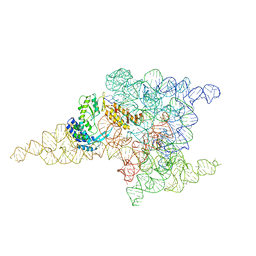 | | Structure of a group II intron retroelement prior to DNA integration | | Descriptor: | MAGNESIUM ION, Maturase reverse transcriptase, SODIUM ION, ... | | Authors: | Haack, D, Yan, X, Zhang, C, Hingey, J, Lyumkis, D, Baker, T.S, Toor, N. | | Deposit date: | 2018-09-05 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Cell, 178, 2019
|
|
4NCO
 
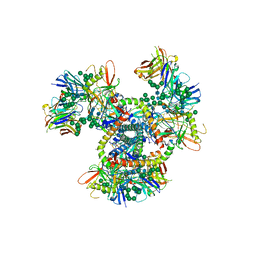 | | Crystal Structure of the BG505 SOSIP gp140 HIV-1 Env trimer in Complex with the Broadly Neutralizing Fab PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIP gp120, BG505 SOSIP gp41, ... | | Authors: | Julien, J.-P, Stanfield, R.L, Lyumkis, D, Ward, A.B, Wilson, I.A. | | Deposit date: | 2013-10-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.7 Å) | | Cite: | Crystal structure of a soluble cleaved HIV-1 envelope trimer.
Science, 342, 2013
|
|
7UT1
 
 | |
7USF
 
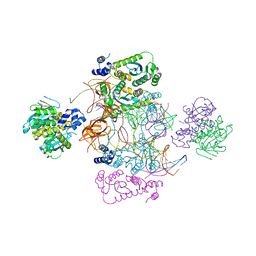 | |
6E9F
 
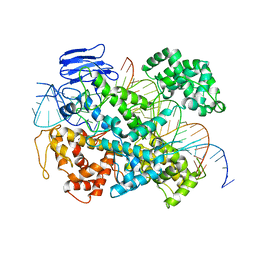 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
6E9E
 
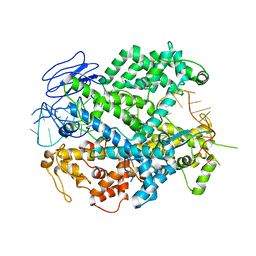 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
6E9D
 
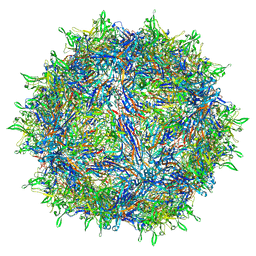 | | Sub-2 Angstrom Ewald Curvature-Corrected Single-Particle Cryo-EM Reconstruction of AAV-2 L336C | | Descriptor: | Capsid protein VP1 | | Authors: | Tan, Y.Z, Aiyer, S, Mietzsch, M, Hull, J.A, McKenna, R, Baker, T.S, Agbandje-McKenna, M, Lyumkis, D. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Sub-2 angstrom Ewald curvature corrected structure of an AAV2 capsid variant.
Nat Commun, 9, 2018
|
|
6MEC
 
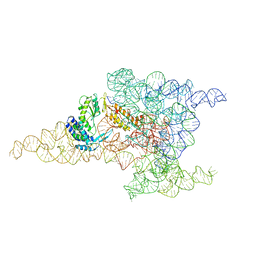 | | Structure of a group II intron retroelement after DNA integration | | Descriptor: | MAGNESIUM ION, Maturase reverse transcriptase, SODIUM ION, ... | | Authors: | Haack, D, Yan, X, Zhang, C, Hingey, J, Lyumkis, D, Baker, T.S, Toor, N. | | Deposit date: | 2018-09-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM Structures of a Group II Intron Reverse Splicing into DNA.
Cell, 178, 2019
|
|
8SY5
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY7
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-STP base pair in the active site | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
5EBZ
 
 | | Crystal structure of human IKK1 | | Descriptor: | 2,3-di-O-sulfo-alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-6)-2,4-di-O-sulfo-alpha-D-glucopyranose, 2-azanyl-5-phenyl-3-(4-sulfamoylphenyl)benzamide, Inhibitor of nuclear factor kappa-B kinase subunit alpha, ... | | Authors: | Polley, S, Passos, D, Huang, D, Biswas, T, Verma, I, Lyumkis, D, Ghosh, G. | | Deposit date: | 2015-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural Basis for the Activation of IKK1/ alpha.
Cell Rep, 17, 2016
|
|
