6VGI
 
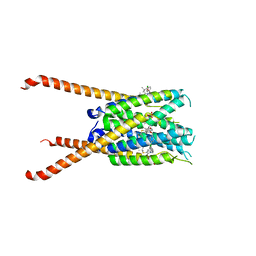 | | Crystal Structures of FLAP bound to MK-866 | | Descriptor: | 3-[3-(tert-butylsulfanyl)-1-[(4-chlorophenyl)methyl]-5-(propan-2-yl)-1H-indol-2-yl]-2,2-dimethylpropanoic acid, 5-lipoxygenase-activating protein, SULFATE ION | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VGC
 
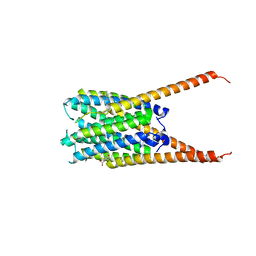 | | Crystal Structures of FLAP bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, 5-lipoxygenase-activating protein, CALCIUM ION, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-07 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
6VL4
 
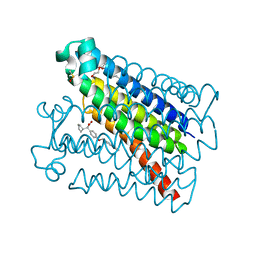 | | Crystal Structure of mPGES-1 bound to DG-031 | | Descriptor: | (2R)-cyclopentyl{4-[(quinolin-2-yl)methoxy]phenyl}acetic acid, Prostaglandin E synthase, TETRAETHYLENE GLYCOL, ... | | Authors: | Ho, J.D, Lee, M.R, Rauch, C.T, Aznavour, K, Park, J.S, Luz, J.G, Antonysamy, S, Condon, B, Maletic, M, Zhang, A, Hickey, M.J, Hughes, N.E, Chandrasekhar, S, Sloan, A.V, Gooding, K, Harvey, A, Yu, X.P, Kahl, S.D, Norman, B.H. | | Deposit date: | 2020-01-22 | | Release date: | 2020-12-02 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based, multi-targeted drug discovery approach to eicosanoid inhibition: Dual inhibitors of mPGES-1 and 5-lipoxygenase activating protein (FLAP).
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
5KW2
 
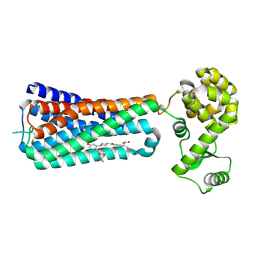 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
2A3X
 
 | | Decameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy- 2-methyl-1,3-dioxane]- 5-yloxycarbonyl}-piperazine | | Descriptor: | BIS-1,2-{[(Z)-2CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBONYL}-PIPERAZINE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
2A3W
 
 | | Decameric structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2-carboxy-2-methyl-1,3-dioxane]-5-yloxycarbamoyl}-ethane | | Descriptor: | BIS-1,2-{[(Z)-2-CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBAMOYL}-ETHANE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
2A3Y
 
 | | Pentameric crystal structure of human serum amyloid P-component bound to Bis-1,2-{[(Z)-2carboxy-2-methyl-1,3-dioxane]-5-yloxycarbamoyl}-ethane. | | Descriptor: | BIS-1,2-{[(Z)-2-CARBOXY-2-METHYL-1,3-DIOXANE]-5-YLOXYCARBAMOYL}-ETHANE, CALCIUM ION, Serum amyloid P-component | | Authors: | Ho, J.G, Kitov, P.I, Paszkiewicz, E, Sadowska, J, Bundle, D.R, Ng, K.K. | | Deposit date: | 2005-06-27 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-assisted Aggregation of Proteins: DIMERIZATION OF SERUM AMYLOID P COMPONENT BY BIVALENT LIGANDS.
J.Biol.Chem., 280, 2005
|
|
3GD8
 
 | | Crystal Structure of Human Aquaporin 4 at 1.8 and its Mechanism of Conductance | | Descriptor: | Aquaporin-4, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Ho, J.D, Yeh, R, Sandstrom, A, Chorny, I, Harries, W.E.C, Robbins, R.A, Miercke, L.J.W, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2G7C
 
 | | Clostridium difficile Toxin A Fragment Bound to aGal(1,3)bGal(1,4)bGlcNAc | | Descriptor: | GLYCEROL, Toxin A, alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Greco, A, Ho, J.G.S, Lin, S.J, Palcic, M.M, Rupnik, M, Ng, K.K.S. | | Deposit date: | 2006-02-28 | | Release date: | 2006-04-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carbohydrate recognition by Clostridium difficile toxin A.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1GNE
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF GLUTATHIONE S-TRANSFERASE OF SCHISTOSOMA JAPONICUM FUSED WITH A CONSERVED NEUTRALIZING EPITOPE ON GP41 OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE | | Authors: | Lim, K, Ho, J.X, Keeling, K, Gilliland, G.L, Ji, X, Ruker, F, Carter, D.C. | | Deposit date: | 1994-06-16 | | Release date: | 1994-11-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV.
Protein Sci., 3, 1994
|
|
1N5U
 
 | | X-RAY STUDY OF HUMAN SERUM ALBUMIN COMPLEXED WITH HEME | | Descriptor: | MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SERUM ALBUMIN | | Authors: | Wardell, M, Wang, Z, Ho, J.X, Robert, J, Ruker, F, Ruble, J, Carter, D.C. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Atomic Structure of Human Methemalbumin at 1.9 A
Biochem.Biophys.Res.Commun., 291, 2002
|
|
4L9K
 
 | | X-ray study of human serum albumin complexed with camptothecin | | Descriptor: | (2S)-2-hydroxy-2-[8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]butanoic acid, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2013-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LB2
 
 | | X-ray study of human serum albumin complexed with idarubicin | | Descriptor: | IDARUBICIN, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LB9
 
 | | X-ray study of human serum albumin complexed with etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol-5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, MYRISTIC ACID, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L8U
 
 | | X-ray study of human serum albumin complexed with 9 amino camptothecin | | Descriptor: | (2S)-2-[1-amino-8-(hydroxymethyl)-9-oxo-9,11-dihydroindolizino[1,2-b]quinolin-7-yl]-2-hydroxybutanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-17 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4LA0
 
 | | X-ray study of human serum albumin complexed with bicalutamide | | Descriptor: | R-BICALUTAMIDE, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
4L9Q
 
 | | X-ray study of human serum albumin complexed with teniposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-(thiophen-2-ylmethylidene)-beta-D-glucopyranoside, SERUM ALBUMIN | | Authors: | Wang, Z, Ho, J.X, Ruble, J, Rose, J.P, Carter, D.C. | | Deposit date: | 2013-06-18 | | Release date: | 2013-07-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of several clinically important oncology drugs in complex with human serum albumin.
Biochim.Biophys.Acta, 1830, 2013
|
|
1O5E
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE, Serine protease hepsin | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5D
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, Coagulation factor VII, Tissue factor | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5A
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 3-{5-[AMINO(IMINIO)METHYL]-1H-INDOL-2-YL}-1,1'-BIPHENYL-2-OLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5G
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CALCIUM ION, Hirudin IIIB', ... | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
1O5B
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5C
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, CITRIC ACID, Urokinase-type plasminogen activator | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA)
J.Mol.Biol., 344, 2004
|
|
1O5F
 
 | | Dissecting and Designing Inhibitor Selectivity Determinants at the S1 site Using an Artificial Ala190 Protease (Ala190 uPA) | | Descriptor: | 2-{5-[AMINO(IMINIO)METHYL]-6-FLUORO-1H-BENZIMIDAZOL-2-YL}-6-[(2-METHYLCYCLOHEXYL)OXY]BENZENOLATE, Serine protease hepsin | | Authors: | Katz, B.A, Luong, C, Ho, J.D, Somoza, J.R, Gjerstad, E, Tang, J, Williams, S.R, Verner, E, Mackman, R.L, Young, W.B, Sprengeler, P.A, Chan, H, Mortara, K, Janc, J.W, McGrath, M.E. | | Deposit date: | 2003-09-09 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dissecting and designing inhibitor selectivity determinants at the S1 site using an artificial Ala190 protease (Ala190 uPA).
J.Mol.Biol., 344, 2004
|
|
2FFX
 
 | | Structure of Human Ferritin L. Chain | | Descriptor: | CADMIUM ION, SULFATE ION, ferritin light chain | | Authors: | Wang, Z.M, Li, C, Ellenburg, M.P, Soitsman, E.M, Ruble, J.R, Wright, B.S, Ho, J.X, Carter, D.C. | | Deposit date: | 2005-12-20 | | Release date: | 2006-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human ferritin L chain
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
