8JKS
 
 | |
8JKO
 
 | |
8JKL
 
 | | IRF4 DNA-binding domain bound to an DNA containing GATA motif | | Descriptor: | GATA-Forward, GATA-Reverse, Interferon regulatory factor 4 | | Authors: | Wang, G, Feng, X, Ding, J. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular basis for the functional roles of the multimorphic T95R mutation of IRF4 causing human autosomal dominant combined immunodeficiency.
Structure, 31, 2023
|
|
8JKQ
 
 | |
7BTA
 
 | | Crystal structure of Rheb D60K mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-03-31 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTC
 
 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
7BTD
 
 | | Crystal structure of Rheb Y35N mutant bound to GppNHp | | Descriptor: | GTP-binding protein Rheb, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
4X9Z
 
 | | Dimeric conotoxin alphaD-GeXXA | | Descriptor: | alphaD-conotoxin GeXXA from the venom of Conus generalis | | Authors: | Xu, S, Zhang, T, Kompella, S, Adams, D, Ding, J, Wang, C. | | Deposit date: | 2014-12-12 | | Release date: | 2015-12-02 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conotoxin alpha D-GeXXA utilizes a novel strategy to antagonize nicotinic acetylcholine receptors
Sci Rep, 5, 2015
|
|
4XPM
 
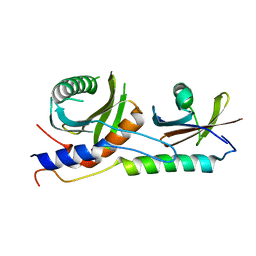 | | Crystal structure of EGO-TC | | Descriptor: | Protein MEH1, Protein SLM4, Uncharacterized protein YCR075W-A | | Authors: | Powis, K, Zhang, T, De Virgilio, C, Ding, J. | | Deposit date: | 2015-01-17 | | Release date: | 2015-08-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling.
Cell Res., 25, 2015
|
|
8GS5
 
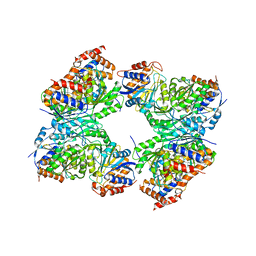 | | Crystal structure of a constitutively active mutant of human IDH3 holoenzyme in apo form | | Descriptor: | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.486 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRG
 
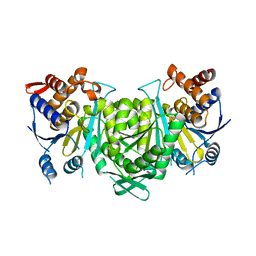 | |
8GRH
 
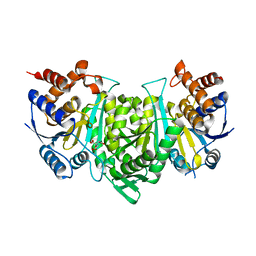 | | Crystal structure of a constitutively active mutant of the alpha gamma heterodimer of human IDH3 in complex with CIT | | Descriptor: | CITRIC ACID, Human IDH3 alpha subunit, Isocitrate dehydrogenase [NAD] subunit gamma, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRU
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ICT, NAD and Ca | | Descriptor: | CALCIUM ION, Human IDH3 alpha subunit, ISOCITRIC ACID, ... | | Authors: | Sun, P, Chen, X, Ding, J. | | Deposit date: | 2022-09-02 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.847 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
8GRB
 
 | |
8GRD
 
 | | Crystal structure of a constitutively active mutant of the alpha beta heterodimer of human IDH3 in complex with ADP and Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial, ... | | Authors: | Chen, X, Sun, P, Ding, J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structures of a constitutively active mutant of human IDH3 reveal new insights into the mechanisms of allosteric activation and the catalytic reaction.
J.Biol.Chem., 298, 2022
|
|
4QTK
 
 | | Complex of WOPR domain of Wor1 in Candida albicans with the 17bp dsDNA | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*TP*AP*AP*AP*CP*TP*TP*TP*TP*TP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*AP*AP*AP*AP*AP*GP*TP*TP*TP*AP*AP*CP*TP*T)-3'), White-opaque regulator 1 | | Authors: | Zhang, S, Zhang, T, Ding, J. | | Deposit date: | 2014-07-08 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal structure of the WOPR-DNA complex and implications for Wor1 function in white-opaque switching of Candida albicans.
Cell Res., 24, 2014
|
|
4QXA
 
 | | Crystal structure of the Rab9A-RUTBC2 RBD complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-9A, ... | | Authors: | Zhang, Z, Wang, S, Ding, J. | | Deposit date: | 2014-07-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Rab9A-RUTBC2 RBD complex reveals the molecular basis for the binding specificity of Rab9A with RUTBC2.
Structure, 22, 2014
|
|
4RL2
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RM5
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4RL0
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | Descriptor: | (2R,5S)-5-[(carbamoyloxy)methyl]-2-[(R)-carboxy{[(2Z)-2-(furan-2-yl)-2-(methoxyimino)acetyl]amino}methyl]-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
4ROV
 
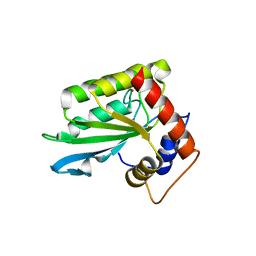 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
4ROW
 
 | | The crystal structure of novel APOBEC3G CD2 head-to-tail dimer suggests the binding mode of full-length APOBEC3G to HIV-1 ssDNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Lu, X, Zhang, T, Xu, Z, Liu, S, Zhao, B, Lan, W, Wang, C, Ding, J, Cao, C. | | Deposit date: | 2014-10-29 | | Release date: | 2014-12-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of DNA cytidine deaminase ABOBEC3G catalytic deamination domain suggests a binding mode of full-length enzyme to single-stranded DNA
J.Biol.Chem., 290, 2015
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
