6SAF
 
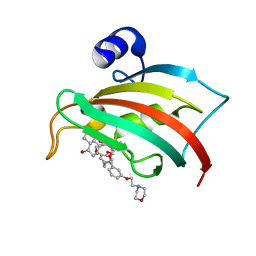 | | The Fk1 domain of FKBP51 in complex with (S)-(R)-3-(3,4-dimethoxyphenyl)-1-(3-(2-morpholinoethoxy)phenyl)propyl 1-((1R,4aR,8aR)-4-oxodecahydronaphthalene-1-carbonyl)piperidine-2-carboxylate | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, [(1~{R})-3-(3,4-dimethoxyphenyl)-1-[3-(2-morpholin-4-ylethoxy)phenyl]propyl] (2~{S})-1-[[(1~{R},4~{a}~{R},8~{a}~{R})-4-oxidanylidene-2,3,4~{a},5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-1-yl]carbonyl]piperidine-2-carboxylate | | Authors: | Feng, X, Sippel, C, Knaup, F, Bracher, A, Staibano, S, Romano, M.F, Hausch, F. | | Deposit date: | 2019-07-16 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Novel Decalin-Based Bicyclic Scaffold for FKBP51-Selective Ligands.
J.Med.Chem., 63, 2020
|
|
8DTP
 
 | | Close state of T4 bacteriophage gp41 hexamer bound with single strand DNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DnaB-like replicative helicase, MAGNESIUM ION, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DWJ
 
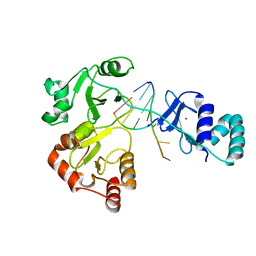 | |
8DUE
 
 | |
8DVF
 
 | | T4 Bacteriophage primosome with single strand DNA, state 1 | | Descriptor: | DNA (5'-D(P*GP*GP*CP*TP*G)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DW6
 
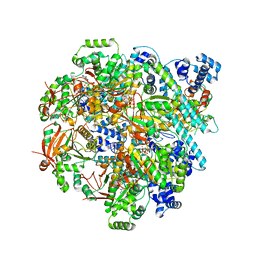 | | T4 bacteriophage primosome with single-strand DNA, State 3 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (70-MER), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-31 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DUO
 
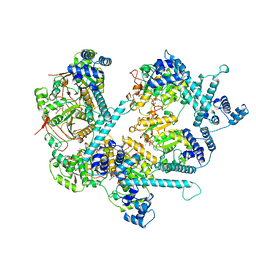 | | DNA-free T4 Bacteriophage gp41 hexamer | | Descriptor: | DnaB-like replicative helicase, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-27 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8DVI
 
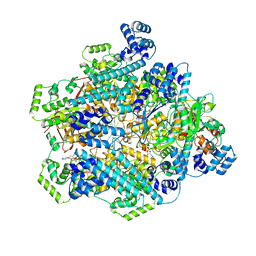 | | T4 bacteriophage primosome with single strand DNA, State 2 | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (70-MER), DNA primase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2022-07-29 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
7T2R
 
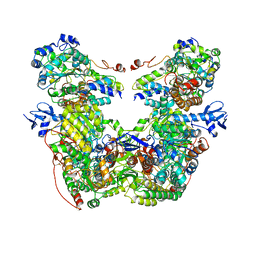 | |
7T30
 
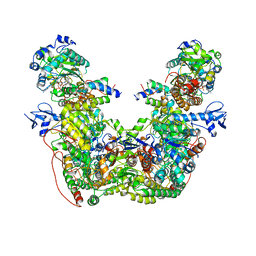 | |
8GAO
 
 | | bacteriophage T4 stalled primosome with mutant gp41-E227Q | | Descriptor: | DNA (70-mer), DNA primase, DnaB-like replicative helicase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
8G0Z
 
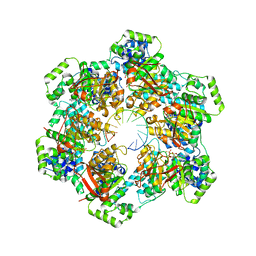 | |
5DIT
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2015-10-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
7KOE
 
 | | Electron bifurcating flavoprotein Fix/EtfABCX | | Descriptor: | Electron transfer flavoprotein, alpha subunit, beta subunit, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2020-11-08 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MCA
 
 | |
4ONC
 
 | | Crystal Structure of Mycobacterium Tuberculosis Decaprenyl Diphosphate Synthase in Complex with BPH-640 | | Descriptor: | Decaprenyl diphosphate synthase, [hydroxy(1,1':3',1''-terphenyl-3-yl)methanediyl]bis(phosphonic acid) | | Authors: | Feng, X, Chan, H.C, Ko, T.P. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and Inhibition of Tuberculosinol Synthase and Decaprenyl Diphosphate Synthase from Mycobacterium tuberculosis
J.Am.Chem.Soc., 136, 2014
|
|
6LJH
 
 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+ | | Descriptor: | Alcohol dehydrogenase 1, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Feng, X, Fan, S, Li, M, Zou, S, Lv, G, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2019-12-16 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua in complex with NAD+
To Be Published
|
|
7CYI
 
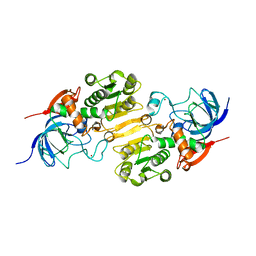 | | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua | | Descriptor: | Alcohol dehydrogenase 1, ZINC ION | | Authors: | Feng, X, Fan, S, Lv, G, Li, M, Wu, G, Jin, Y, Yang, Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of Alcohol dehydrogenase 1 from Artemisia annua
To Be Published
|
|
4Z2Z
 
 | |
7UQJ
 
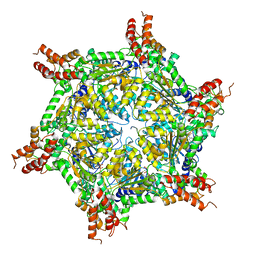 | | Cryo-EM structure of the S. cerevisiae chromatin remodeler Yta7 hexamer bound to ATPgS and histone H3 tail in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase histone chaperone YTA7, Histone H3, ... | | Authors: | Wang, F, Feng, X, Li, H. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-01 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Saccharomyces cerevisiae Yta7 ATPase hexamer contains a unique bromodomain tier that functions in nucleosome disassembly.
J.Biol.Chem., 299, 2022
|
|
7UQK
 
 | |
7UQI
 
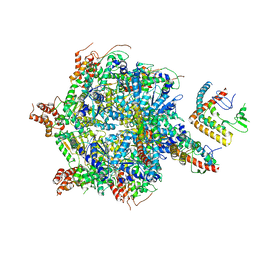 | |
7X9Q
 
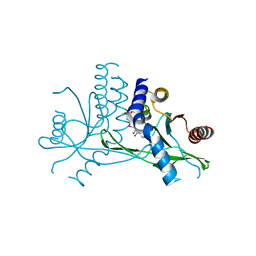 | | Crystal structure of human STING complexed with compound BSP16 | | Descriptor: | (2R)-4-(5,6-dimethoxy-1-benzoselenophen-2-yl)-2-ethyl-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Selenium-Containing STING Agonists as Orally Available Antitumor Agents.
J.Med.Chem., 65, 2022
|
|
7X9P
 
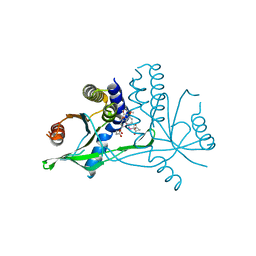 | | Crystal structure of human STING complexed with compound BSP17 | | Descriptor: | 4-[6-methoxy-5-[3-[[6-methoxy-2-(4-oxidanyl-4-oxidanylidene-butanoyl)-1-benzoselenophen-5-yl]oxy]propoxy]-1-benzoselenophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Pan, L, Guan, X, Feng, X, Li, Z, Bian, J. | | Deposit date: | 2022-03-15 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Selenium-containing STING agonists as orally available anti tumor agents
To be published
|
|
5DIV
 
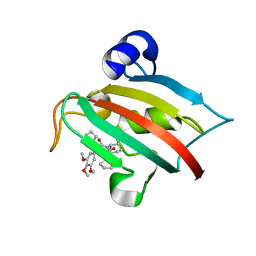 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (S)-N-(1-carbamoylcyclopentyl)-1-((S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carboxamide | | Descriptor: | (2S)-N-(1-carbamoylcyclopentyl)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Gaali, S, Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-23 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rapid, Structure-Based Exploration of Pipecolic Acid Amides as Novel Selective Antagonists of the FK506-Binding Protein 51.
J.Med.Chem., 59, 2016
|
|
