2KKU
 
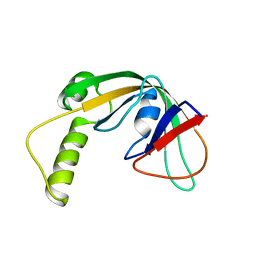 | | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351 | | Descriptor: | Uncharacterized protein | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Rumpel, S, Semest, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-06-29 | | Release date: | 2009-07-14 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of protein af2351 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium target AtT9/Ontario Center for Structural Proteomics Target af2351
To be Published
|
|
2KKX
 
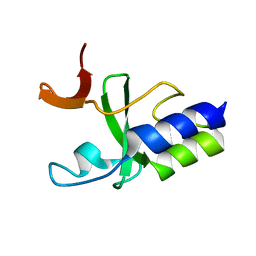 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
6FAE
 
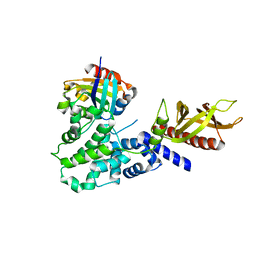 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | Authors: | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
6FWZ
 
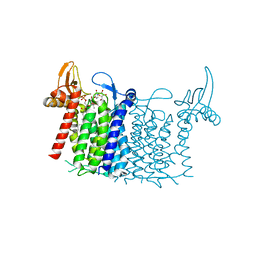 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-07 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT7
 
 | | Crystal structure of CLK3 in complex with compound 8a | | Descriptor: | 1,2-ETHANEDIOL, 3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, Dual specificity protein kinase CLK3, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6FT9
 
 | | Crystal structure of CLK1 in complex with inhibitor 16 | | Descriptor: | 2-bromanyl-3-phenyl-1~{H}-pyrrolo[3,4-g]indol-8-one, BROMIDE ION, Dual specificity protein kinase CLK1, ... | | Authors: | Chaikuad, A, Walter, A, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Molecular structures of cdc2-like kinases in complex with a new inhibitor chemotype.
PLoS ONE, 13, 2018
|
|
6G5P
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment FM009493b 2,3-Dimethoxy-2,3-dimethyl-2,3-dihydro-1,4-benzodioxin-6-amine | | Descriptor: | (2~{R},3~{R})-2,3-dimethoxy-2,3-dimethyl-1,4-benzodioxin-6-amine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
6FM9
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
6FU5
 
 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP18 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[5-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-2-methoxy-phenyl]propane-1-sulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Small molecule inhibitors reveal an indispensable scaffolding role of RIPK2 in NOD2 signaling.
EMBO J., 37, 2018
|
|
6G37
 
 | | Crystal structure of haspin in complex with 5-fluorotubercidin | | Descriptor: | 5-fluorotubercidin, PHOSPHATE ION, Serine/threonine-protein kinase haspin | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G3A
 
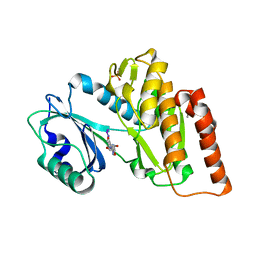 | | Crystal structure of haspin F605T mutant in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6GES
 
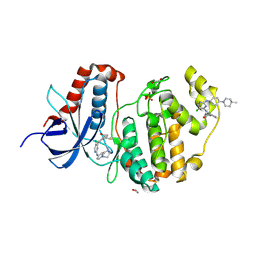 | | Crystal structure of ERK1 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 3, N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]phenyl}propanamide, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6G33
 
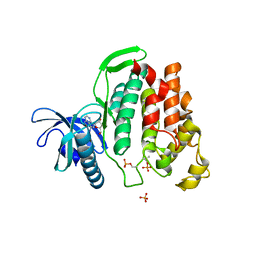 | | Crystal structure of CLK1 in complex with 5-iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Dual specificity protein kinase CLK1, IODIDE ION, ... | | Authors: | Heroven, C, Chaikuad, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-24 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Halogen-Aromatic pi Interactions Modulate Inhibitor Residence Times.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6G54
 
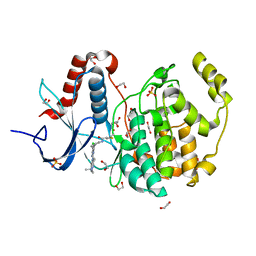 | | Crystal structure of ERK2 covalently bound to SM1-71 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 1, ... | | Authors: | Chaikuad, A, Suman, R, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-29 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Leveraging Compound Promiscuity to Identify Targetable Cysteines within the Kinome.
Cell Chem Biol, 26, 2019
|
|
6G0O
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G0P
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated E2F1 peptide (K117ac/K120ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcription factor E2F1 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, Sorrell, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
6G5N
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment XS039818e 1-(3-Phenyl-1,2,4-oxadiazol-5-yl)methanamine | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
6GL9
 
 | | Crystal structure of JAK3 in complex with Compound 10 (FM475) | | Descriptor: | (~{E})-3-[3-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)phenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6GI6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6GIP
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 2, 5-dimethyl core. | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dimethyl-6-quinolin-4-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mathea, S, Chen, Z, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6GLA
 
 | | Crystal structure of JAK3 in complex with Compound 11 (FM481) | | Descriptor: | (~{E})-3-[5-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)furan-2-yl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
6GY5
 
 | | Crystal structure of the kelch domain of human KLHL20 in complex with DAPK1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Chen, Z, Hozjan, V, Strain-Damerell, C, Williams, E, Wang, D, Cooper, C.D.O, Sanvitale, C.E, Fairhead, M, Carpenter, E.P, Pike, A.C.W, Krojer, T, Srikannathasan, V, Sorrell, F, Johansson, C, Mathea, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase.
Structure, 27, 2019
|
|
