7RMA
 
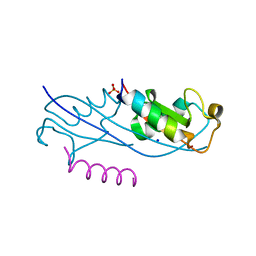 | | Structure of the fourth UIM (Ubiquitin Interacting Motif) of ANKRD13D in complex with a high affinity UbV (Ubiquitin Variant) | | Descriptor: | Ankyrin repeat domain-containing protein 13D, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Manczyk, N, Veggiani, G, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-07-27 | | Release date: | 2022-05-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Panel of Engineered Ubiquitin Variants Targeting the Family of Human Ubiquitin Interacting Motifs.
Acs Chem.Biol., 17, 2022
|
|
6UBH
 
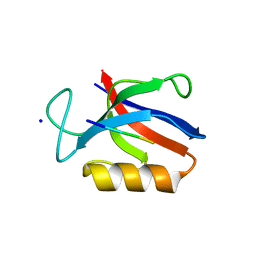 | | Structure of the MM7 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | Erbin, SODIUM ION, peptide | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-09-11 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
6ML1
 
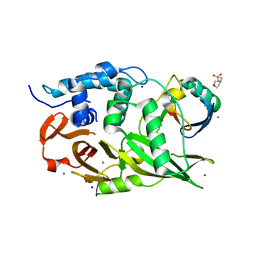 | | Structure of the USP15 deubiquitinase domain in complex with an affinity-matured inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6Q0U
 
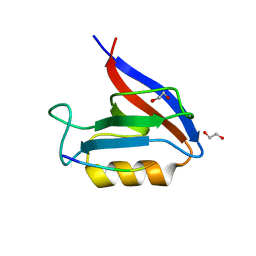 | | Structure of the Erbin PDZ variant E-6a with a high-affinity C-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q0M
 
 | | Structure of Erbin PDZ derivative E-14 with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
6Q0N
 
 | | Structure of the Erbin PDB domain in complex with a high-affinity peptide | | Descriptor: | Erbin, peptide | | Authors: | Singer, A.U, Teyra, J, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2019-08-02 | | Release date: | 2019-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Comprehensive analysis of all evolutionary paths between two divergent PDZ domain specificities.
Protein Sci., 29, 2020
|
|
7LUL
 
 | | Structure of the MM2 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
6DJ9
 
 | | Structure of the USP15 DUSP domain in complex with a high-affinity Ubiquitin Variant (UbV) | | Descriptor: | Ubiquitin Variant UbV 15.D, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-05-24 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
8T7G
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
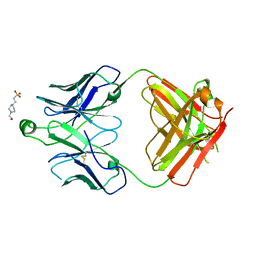 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8CX9
 
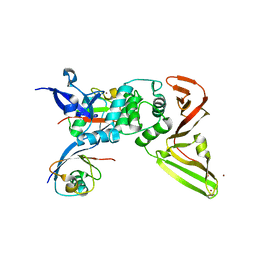 | | Structure of the SARS-COV2 PLpro (C111S) in complex with a dimeric Ubv that inhibits activity by an unusual allosteric mechanism | | Descriptor: | BROMIDE ION, CHLORIDE ION, Papain-like protease nsp3, ... | | Authors: | Singer, A.U, Slater, C.L, Patel, A, Russel, R, Mark, B.L, Sidhu, S.S. | | Deposit date: | 2022-05-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site.
Plos Pathog., 18, 2022
|
|
8T7I
 
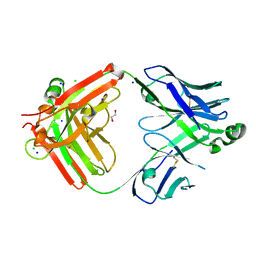 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TS5
 
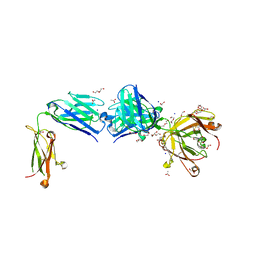 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRT
 
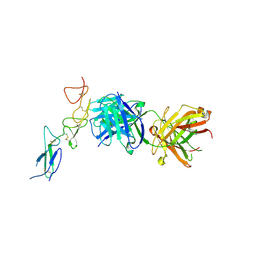 | | Structure of the EphA2 CRD bound to FabS1CE_C1, monoclinic form | | Descriptor: | CHLORIDE ION, Ephrin type-A receptor 2, S1CE variant of Fab C1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TRS
 
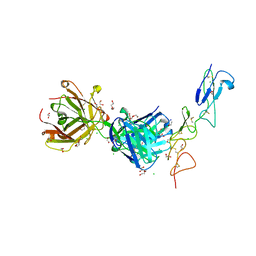 | | Structure of the EphA2 CRD bound to FabS1CE_C1, trigonal form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
6CRN
 
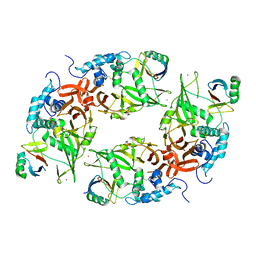 | | Structure of the USP15 deubiquitinase domain in complex with a high-affinity first-generation Ubv | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15, Ubiquitin variant 15.2, ZINC ION | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-19 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6CPM
 
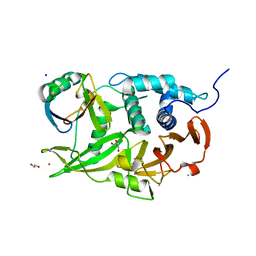 | | Structure of the USP15 deubiquitinase domain in complex with a third-generation inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
4FCG
 
 | | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Zimmerman, M.D, Minor, W, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the leucine-rich repeat domain of the type III effector XCV3220 (XopL)
To be Published
|
|
4FC9
 
 | | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, uncharacterized protein | | Authors: | Singer, A.U, Xu, X, Cui, H, Tan, K, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-24 | | Release date: | 2012-06-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminal domain of the type III effector Xcv3220 (XopL)
To be Published
|
|
3SVI
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited thermolysin digestion | | Descriptor: | CHLORIDE ION, SODIUM ION, SULFATE ION, ... | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-12 | | Release date: | 2011-08-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analysis of HopPmaL Reveals the Presence of a Second Adaptor Domain Common to the HopAB Family of Pseudomonas syringae Type III Effectors.
Biochemistry, 51, 2012
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | Descriptor: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
3T7Y
 
 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
1S28
 
 | | Crystal Structure of AvrPphF ORF1, the Chaperone for the Type III Effector AvrPphF ORF2 from P. syringae | | Descriptor: | ORF1, SULFATE ION | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.L, Sondek, J. | | Deposit date: | 2004-01-08 | | Release date: | 2004-09-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
2QH1
 
 | | Structure of TA289, a CBS-rubredoxin-like protein, in its Fe+2-bound state | | Descriptor: | FE (II) ION, Hypothetical protein Ta0289 | | Authors: | Singer, A.U, Proudfoot, M, Brown, G, Xu, L, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-06-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain.
J.Mol.Biol., 375, 2008
|
|
1S21
 
 | | Crystal Structure of AvrPphF ORF2, A Type III Effector from P. syringae | | Descriptor: | ORF2 | | Authors: | Singer, A.U, Desveaux, D, Betts, L, Chang, J.H, Nimchuk, Z, Grant, S.R, Dangl, J.K, Sondek, J. | | Deposit date: | 2004-01-07 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of the Type III Effector Protein AvrPphF and Its Chaperone Reveal Residues Required for Plant Pathogenesis
Structure, 12, 2004
|
|
