5KXA
 
 | | Selective Inhibition of Autotaxin is Effective in Mouse Models of Liver Fibrosis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[6-chloranyl-2-cyclopropyl-1-(1-ethylpyrazol-4-yl)-7-fluoranyl-indol-3-yl]sulfanyl-2-fluoranyl-benzoic acid, CALCIUM ION, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2016-07-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Selective Inhibition of Autotaxin Is Efficacious in Mouse Models of Liver Fibrosis.
J. Pharmacol. Exp. Ther., 360, 2017
|
|
5L01
 
 | | Tryptophan 5-hydroxylase in complex with inhibitor (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid | | Descriptor: | (3~{S})-8-[2-azanyl-6-[(1~{R})-1-(4-chloranyl-2-phenyl-phenyl)-2,2,2-tris(fluoranyl)ethoxy]pyrimidin-4-yl]-2,8-diazaspiro[4.5]decane-3-carboxylic acid, FE (III) ION, Tryptophan 5-hydroxylase 1 | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-07-26 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6MO5
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
1JKI
 
 | | myo-Inositol-1-phosphate Synthase Complexed with an Inhibitor, 2-deoxy-glucitol-6-phosphate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-DEOXY-GLUCITOL-6-PHOSPHATE, AMMONIUM ION, ... | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1JKF
 
 | | Holo 1L-myo-inositol-1-phosphate Synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, myo-inositol-1-phosphate synthase | | Authors: | Stein, A.J, Geiger, J.H. | | Deposit date: | 2001-07-12 | | Release date: | 2002-04-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure and mechanism of 1-L-myo-inositol- 1-phosphate synthase
J.Biol.Chem., 277, 2002
|
|
1YVP
 
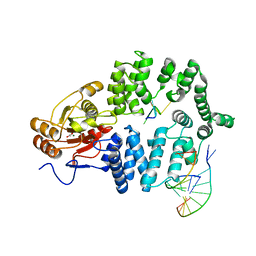 | | Ro autoantigen complexed with RNAs | | Descriptor: | 60-kDa SS-A/Ro ribonucleoprotein, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Stein, A.J, Fuchs, G, Fu, C, Wolin, S.L, Reinisch, K.M. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into RNA Quality Control: The Ro Autoantigen Binds Misfolded RNAs via Its Central Cavity
Cell(Cambridge,Mass.), 121, 2005
|
|
1YVR
 
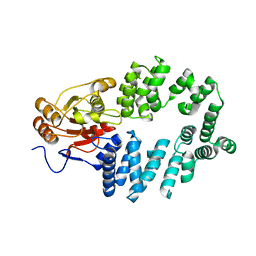 | | Ro autoantigen | | Descriptor: | 60-kDa SS-A/Ro ribonucleoprotein | | Authors: | Stein, A.J, Fuchs, G, Fu, C, Wolin, S.L, Reinisch, K.M. | | Deposit date: | 2005-02-16 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into RNA Quality Control: The Ro Autoantigen Binds Misfolded RNAs via Its Central Cavity
Cell(Cambridge,Mass.), 121, 2005
|
|
7OND
 
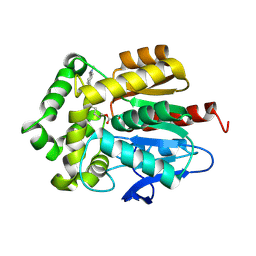 | |
7OO4
 
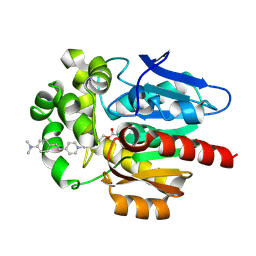 | |
4ZG6
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZG9
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(11aS)-6-(4-fluorobenzyl)-1,3-dioxo-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indol-2(3H)-yl]propanoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZGA
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (11aS)-6-(4-fluorobenzyl)-5,6,11,11a-tetrahydro-1H-imidazo[1',5':1,6]pyrido[3,4-b]indole-1,3(2H)-dione, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
4ZG7
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
7ONP
 
 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONM
 
 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONQ
 
 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONV
 
 | | Carbonic anhydrase II mutant (I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
4J3I
 
 | | X-ray crystal structure of bromodomain complex to 1.24 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
4J1P
 
 | | X-ray crystal structure of bromodomain 2 of human brd2 in complex with rvx208 to 1.08 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain containing 2, ... | | Authors: | Stein, A.J, White, A, Suto, R.K. | | Deposit date: | 2013-02-01 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | RVX-208, an Inducer of ApoA-I in Humans, Is a BET Bromodomain Antagonist.
Plos One, 8, 2013
|
|
5J6D
 
 | | Discovery of acyl guanidine tryptophan hydroxylase-1 inhibitors | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[(N-{[2-(3-methoxyphenoxy)-6-(piperidin-1-yl)phenyl]methyl}carbamimidoyl)carbamoyl]-L-phenylalanine, FE (III) ION, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S. | | Deposit date: | 2016-04-04 | | Release date: | 2016-05-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of acyl guanidine tryptophan hydroxylase-1 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5T31
 
 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5TPG
 
 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | Descriptor: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | Authors: | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
