5O33
 
 | | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Kalirin, ... | | Authors: | Gray, J, Krojer, T, Talon, R, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-05-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A structure of the GEF Kalirin DH1 domain in complex with the small GTPase Rac1
To Be Published
|
|
5QQN
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-010-382-606 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-cyclopropyl-2-(4-{[(5-methyl-1,2-oxazol-3-yl)carbamoyl]amino}-1H-pyrazol-1-yl)acetamide, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQE
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-531-494 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[(3S)-4,4,4-trifluoro-3-hydroxy-3-(5-methylfuran-2-yl)butyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQK
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-359-835 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[2-(diethylamino)ethyl]-1H-indol-5-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQG
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-541-216 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[3-(4-phenylpiperazin-1-yl)propyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-004-412-710 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-1,3-thiazol-2-ylurea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-587-558 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[(imidazo[1,2-a]pyridin-2-yl)methyl]-1H-pyrazol-4-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-325 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(1-benzyl-1H-pyrazol-4-yl)-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-178-994 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-[3-(methoxymethyl)phenyl]-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQL
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-565-301 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-(5-methyl-1,2-oxazol-3-yl)-N'-[2-(phenylsulfonyl)ethyl]urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with Z56880342 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QQH
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-020-096-465 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-methyl-5-({[(5-methyl-1,2-oxazol-3-yl)carbamoyl]amino}methyl)thiophene-2-sulfonamide, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QU9
 
 | | PanDDA analysis group deposition of ground-state model of Kalirin/Rac1 screened against a customized urea fragment library by X-ray Crystallography at the XChem facility of Diamond Light Source beamline I04-1 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
6FAE
 
 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | Authors: | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
6U0X
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS Q123D at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6U0W
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K133M at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-08-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OK8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS K127L at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Jeliazkov, J.R, Robinson, A.C, Berger, J.M, Garcia-Moreno E, B, Gray, J.G. | | Deposit date: | 2019-04-12 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward the computational design of protein crystals with improved resolution.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2VQA
 
 | | Protein-folding location can regulate Mn versus Cu- or Zn-binding. Crystal Structure of MncA. | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SLL1358 PROTEIN | | Authors: | Tottey, S, Waldron, K.J, Firbank, S.J, Reale, B, Bessant, C, Sato, K, Gray, J, Banfield, M.J, Dennison, C, Robinson, N.J. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Protein-Folding Location Can Regulate Manganese-Binding Versus Copper- or Zinc-Binding.
Nature, 455, 2008
|
|
8C7D
 
 | | Structure of the GEF Kalirin DH2 Domain | | Descriptor: | Kalirin | | Authors: | Callens, M.C, Thompson, A.P, Gray, J.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based alignment and analysis of RHO selectivity of RHO-DBL GTPase exchange factors
to be published
|
|
2XJI
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2XJH
 
 | | Structure and Copper-binding Properties of Methanobactins from Methylosinus trichosporium OB3b | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB-OB3B, SODIUM ION | | Authors: | El-Ghazouani, A, Basle, A, Firbank, S.J, Knapp, C.W, Gray, J, Graham, D.W, Dennison, C. | | Deposit date: | 2010-07-06 | | Release date: | 2011-02-02 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
Inorg.Chem., 50, 2011
|
|
2YGI
 
 | | Methanobactin HM1 | | Descriptor: | COPPER (II) ION, METHANOBACTIN HM1 | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2YGJ
 
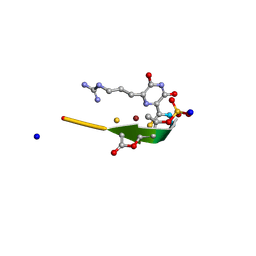 | | Methanobactin MB4 | | Descriptor: | COPPER (II) ION, METHANOBACTIN MB4, SODIUM ION | | Authors: | Ghazouani, A, Basle, A, Firbank, S.J, Gray, J, Dennison, C. | | Deposit date: | 2011-04-18 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Variations in Methanobactin Structure Influences Copper Utilization by Methane-Oxidizing Bacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6GWK
 
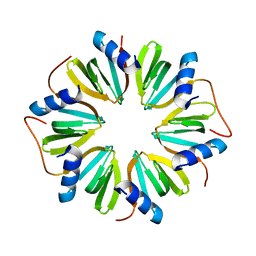 | | The crystal structure of Hfq from Caulobacter crescentus | | Descriptor: | RNA-binding protein Hfq | | Authors: | Santiago-Frangos, A, Frohlich, K.S, Jeliazkov, J.R, Gray, J.R, Luisi, B.F, Woodson, S.A, Hardwick, S.W. | | Deposit date: | 2018-06-25 | | Release date: | 2019-05-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Caulobacter crescentusHfq structure reveals a conserved mechanism of RNA annealing regulation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2H02
 
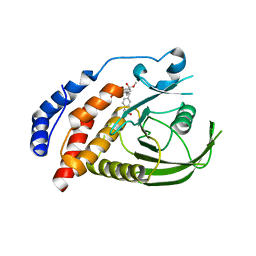 | | Structural studies of protein tyrosine phosphatase beta catalytic domain in complex with inhibitors | | Descriptor: | Protein tyrosine phosphatase, receptor type, B,, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M, Gray, J.L, Peters, K.G, Maier, M.B, Amarasinghe, K.D, Clark, C.M, Nichols, R. | | Deposit date: | 2006-05-13 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of potent, non-peptidic inhibitors of HPTPbeta.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
