5IVY
 
 | |
6UIP
 
 | | DYRK1A Kinase Domain in Complex with a 6-azaindole Derivative, GNF2133. | | Descriptor: | 4-ethyl-N-{4-[1-(oxan-4-yl)-1H-pyrrolo[2,3-c]pyridin-3-yl]pyridin-2-yl}piperazine-1-carboxamide, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133.
J.Med.Chem., 63, 2020
|
|
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
5GWE
 
 | | cytochrome P450 CREJ | | Descriptor: | (4-methylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Dong, S, liu, X, Wang, X, Feng, Y. | | Deposit date: | 2016-09-11 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1YFJ
 
 | | T4Dam in Complex with AdoHcy and 15-mer Oligonucleotide Showing Semi-specific and Specific Contact | | Descriptor: | 5'-D(*TP*CP*AP*CP*AP*GP*GP*AP*TP*CP*CP*TP*GP*TP*G)-3', CALCIUM ION, CHLORIDE ION, ... | | Authors: | Horton, J.R, Liebert, K, Hattman, S, Jeltsch, A, Cheng, X. | | Deposit date: | 2005-01-02 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Transition from Nonspecific to Specific DNA Interactions along the Substrate-Recognition Pathway of Dam Methyltransferase.
Cell(Cambridge,Mass.), 121, 2005
|
|
5UJ3
 
 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed cefotaxime | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-(methoxyimino)acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro -2H-1,3-thiazine-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
5ISL
 
 | |
5U6J
 
 | | Factor VIIa in complex with the inhibitor 3-{[(2R)-17-ethyl-4-methyl-3,12-dioxo-7-[(propan-2-yl)sulfonyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-2-yl]amino}benzamide | | Descriptor: | 3-{[(2R)-17-ethyl-4-methyl-3,12-dioxo-7-[(propan-2-yl)sulfonyl]-13-oxa-4,11-diazatricyclo[14.2.2.1~6,10~]henicosa-1(18),6(21),7,9,16,19-hexaen-2-yl]amino}benzamide, CALCIUM ION, Coagulation factor VII Heavy Chain, ... | | Authors: | Wei, A. | | Deposit date: | 2016-12-08 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Neutral macrocyclic factor VIIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3J2Z
 
 | |
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | Descriptor: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5UJ4
 
 | | Crystal structure of the KPC-2 beta-lactamase complexed with hydrolyzed faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2017-01-16 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Substrate Recognition and Product Release by the Klebsiella pneumoniae Carbapenemase (KPC-2).
J. Med. Chem., 60, 2017
|
|
6J9O
 
 | |
4JVE
 
 | | Co-crystal structure of MDM2 with inhibitor (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid | | Descriptor: | (2R,3E)-2-[(2S,3R,6S)-2,3-bis(4-chlorophenyl)-6-(4-fluorobenzyl)-5-oxomorpholin-4-yl]pent-3-enoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4QO4
 
 | |
3GZT
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7 | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4JV9
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-chlorophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
5UL8
 
 | |
7LTO
 
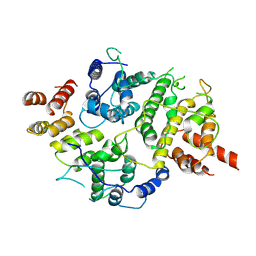 | | Nse5-6 complex | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2MQG
 
 | |
2NB4
 
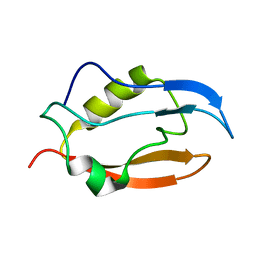 | | Solution structure of Q388A3 PDZ domain | | Descriptor: | Putative uncharacterized protein | | Authors: | Mei, S. | | Deposit date: | 2016-01-24 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Q388A3 PDZ domain from Trypanosoma brucei
J.Struct.Biol., 194, 2016
|
|
2N7S
 
 | | Solution Structure of Leptospiral LigA4 Big Domain | | Descriptor: | Ig-like repeat domain protein 1 | | Authors: | Mei, S. | | Deposit date: | 2015-09-17 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of leptospiral LigA4 Big domain.
Biochem. Biophys. Res. Commun., 467, 2015
|
|
7K36
 
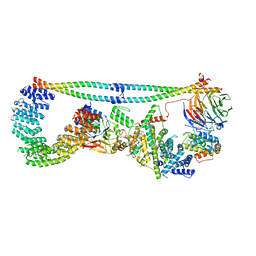 | | Cryo-EM structure of STRIPAK complex | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MANGANESE (II) ION, MOB-like protein phocein, ... | | Authors: | Jeong, B.-C, Bai, X.C. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the Hippo signaling integrator human STRIPAK.
Nat.Struct.Mol.Biol., 28, 2021
|
|
