5KKB
 
 | | Structure of mouse Golgi alpha-1,2-mannosidase IA and Man9GlcNAc2-PA complex | | Descriptor: | 1,4-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, ... | | Authors: | Xiang, Y, Moremen, K.W. | | Deposit date: | 2016-06-21 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Substrate recognition and catalysis by GH47 alpha-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
4U0L
 
 | |
4U0N
 
 | | Structure of the Vibrio cholerae di-nucleotide cyclase (DncV) deletion mutant D-loop | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, N-[4-({[(6S)-2-amino-5-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid | | Authors: | Xiang, Y, Zhu, D.Y. | | Deposit date: | 2014-07-12 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Biochemistry of a Vibrio cholerae Dinucleotide Cyclase Reveals Cyclase Activity Regulation by Folates.
Mol.Cell, 55, 2014
|
|
1P9G
 
 | | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Ecommia ulmoides Oliver at atomic resolution | | Descriptor: | ACETATE ION, EAFP 2 | | Authors: | Xiang, Y, Huang, R.H, Liu, X.Z, Wang, D.C. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Crystal structure of a novel antifungal protein distinct with five disulfide bridges from Eucommia ulmoides Oliver at an atomic resolution.
J.Struct.Biol., 148, 2004
|
|
1T7B
 
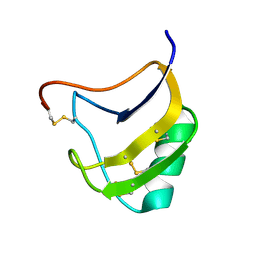 | | Crystal structure of mutant Lys8Gln of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7A
 
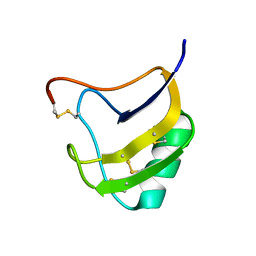 | | Crystal structure of mutant Lys8Asp of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
1T7E
 
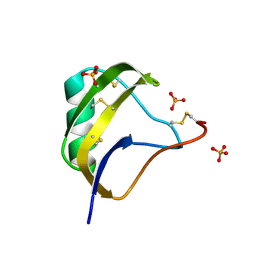 | | Crystal structure of mutant Pro9Ser of scorpion alpha-like neurotoxin BmK M1 from Buthus martensii Karsch | | Descriptor: | Alpha-like neurotoxin BmK-I, PHOSPHATE ION | | Authors: | Xiang, Y, Guan, R.J, He, X.L, Wang, C.G, Wang, M, Zhang, Y, Sundberg, E.J, Wang, D.C. | | Deposit date: | 2004-05-09 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Mechanism Governing Cis and Trans Isomeric States and an Intramolecular Switch for Cis/Trans Isomerization of a Non-proline Peptide Bond Observed in Crystal Structures of Scorpion Toxins
J.Mol.Biol., 341, 2004
|
|
8J1V
 
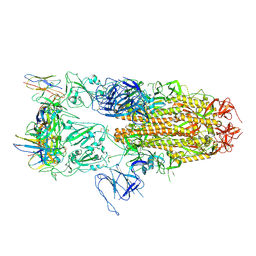 | |
8J1T
 
 | |
8IXL
 
 | | top segment of the bacteriophage M13 mini variant | | Descriptor: | Capsid protein G8P, Tail virion protein G7P, Tail virion protein G9P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
8JWT
 
 | |
8JWX
 
 | |
8JWW
 
 | |
8IXJ
 
 | |
8IXK
 
 | | bottom segment of the bacteriophage M13 mini variant | | Descriptor: | Attachment protein G3P, Capsid protein G8P, Head virion protein G6P | | Authors: | Xiang, Y, Jia, Q. | | Deposit date: | 2023-04-01 | | Release date: | 2023-09-13 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of a bacteriophage M13 mini variant.
Nat Commun, 14, 2023
|
|
3SUC
 
 | |
3R04
 
 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 5-{6-[(trans-4-aminocyclohexyl)amino]pyrazin-2-yl}-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7WC0
 
 | |
5Y66
 
 | | Crystal structure of Pseudomonas fluorescens Kynurenine 3-monooxygenase in complex with L-KYN and Ro61-8048 | | Descriptor: | (2S)-2-amino-4-(2-aminophenyl)-4-oxobutanoic acid, 3,4-dimethoxy-N-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]benzenesulfonamide, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xiang, Y, Gao, J.J, Zhu, D.Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Biochemistry and structural studies of kynurenine 3-monooxygenase reveal allosteric inhibition by Ro 61-8048
FASEB J., 32, 2018
|
|
5Y7A
 
 | |
5Y77
 
 | |
7Y42
 
 | |
3GQ7
 
 | |
3GQ8
 
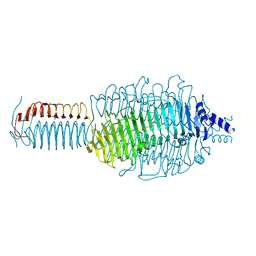 | |
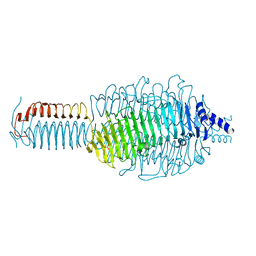
3GQA
 
 | |
