3MKS
 
 | | Crystal Structure of yeast Cdc4/Skp1 in complex with an allosteric inhibitor SCF-I2 | | Descriptor: | 1,1'-binaphthalene-2,2'-dicarboxylic acid, Cell division control protein 4, GLYCEROL, ... | | Authors: | Orlicky, S, Sicheri, F, Tyers, M, Tang, X. | | Deposit date: | 2010-04-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Nat.Biotechnol., 28, 2010
|
|
1NEX
 
 | | Crystal Structure of ScSkp1-ScCdc4-CPD peptide complex | | Descriptor: | CDC4 protein, Centromere DNA-binding protein complex CBF3 subunit D, GLL(TPO)PPQSG | | Authors: | Orlicky, S, Tang, X, Willems, A, Tyers, M, Sicheri, F. | | Deposit date: | 2002-12-12 | | Release date: | 2003-02-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Phosphodependent Substrate Selection and
Orientation by the SCFCdc4 Ubiquitin Ligase
Cell(Cambridge,Mass.), 112, 2003
|
|
5IBK
 
 | | Skp1-F-box in complex with a ubiquitin variant | | Descriptor: | F-box/WD repeat-containing protein 7, Polyubiquitin-B, S-phase kinase-associated protein 1,S-phase kinase-associated protein 1 | | Authors: | Orlicky, S, Sicheri, F. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2P63
 
 | |
4MDK
 
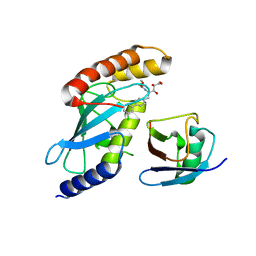 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
3V7D
 
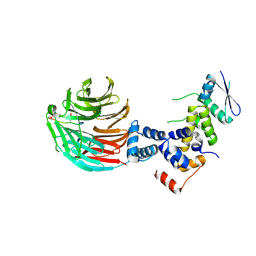 | | Crystal Structure of ScSkp1-ScCdc4-pSic1 peptide complex | | Descriptor: | Cell division control protein 4, Protein SIC1, Suppressor of kinetochore protein 1 | | Authors: | Tang, X, Orlicky, S, Mittag, T, Csizmok, V, Pawson, T, Forman-Kay, J, Sicheri, F, Tyers, M. | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2P64
 
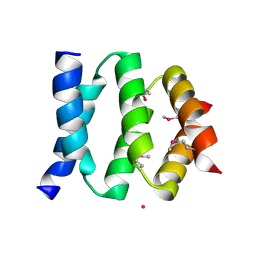 | | D domain of b-TrCP | | Descriptor: | CADMIUM ION, F-box/WD repeat protein 1A | | Authors: | Neculai, D, Orlicky, S, Ceccarelli, D. | | Deposit date: | 2007-03-16 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination.
Cell(Cambridge,Mass.), 129, 2007
|
|
8D6E
 
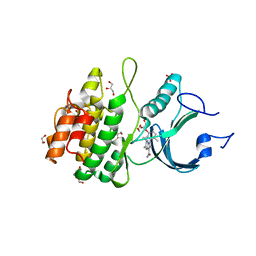 | | Crystal Structure of Human Myt1 Kinase domain Bounded with RP-6306 | | Descriptor: | (1P)-2-amino-1-(3-hydroxy-2,6-dimethylphenyl)-5,6-dimethyl-1H-pyrrolo[2,3-b]pyridine-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6D
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 39 | | Descriptor: | (1P)-2-amino-5-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6F
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with Eph receptor inhibitor / compound 41 | | Descriptor: | (1M)-2-amino-1-(5-hydroxy-2-methylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, SULFATE ION | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
8D6C
 
 | | Crystal Structure of Human Myt1 Kinase domain Bounded with compound 28 | | Descriptor: | (1P)-2-amino-6-bromo-1-(3-hydroxy-2,6-dimethylphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Pau, V.P.T, Mao, D.Y.L, Mader, P, Orlicky, S, Sicheri, F. | | Deposit date: | 2022-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of an Orally Bioavailable and Selective PKMYT1 Inhibitor, RP-6306.
J.Med.Chem., 65, 2022
|
|
6UUO
 
 | | Crystal structure of BRAF kinase domain bound to the PROTAC P4B | | Descriptor: | N-(3-{5-[(1-acetylpiperidin-4-yl)(methyl)amino]-3-(pyrimidin-5-yl)-1H-pyrrolo[3,2-b]pyridin-1-yl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Posternak, G, Kurinov, I, Sicheri, F. | | Deposit date: | 2019-10-30 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Functional characterization of a PROTAC directed against BRAF mutant V600E.
Nat.Chem.Biol., 16, 2020
|
|
7M2K
 
 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
3RZ3
 
 | | Human Cdc34 E2 in complex with CC0651 inhibitor | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Webb, D.R, Sicheri, F. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An allosteric inhibitor of the human cdc34 ubiquitin conjugating enzyme
Cell(Cambridge,Mass.), 145, 2011
|
|
8T9B
 
 | | Structure of the CK variant of Fab F1 (FabC-F1) in complex with the C-terminal FN3 domain of EphA2 | | Descriptor: | CK variant of Fab F1 heavy chain, CK variant of Fab F1 light chain, Ephrin type-A receptor 2 | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Engineered Antigen-binding Fragments for Enhanced Crystallization of Antibody:Antigen Complexes
To be Published
|
|
8TTQ
 
 | |
4O1O
 
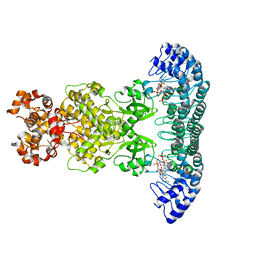 | | Crystal Structure of RNase L in complex with 2-5A | | Descriptor: | Ribonuclease L, [[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3R,4R,5R)-5-(6-aminopurin-9-yl)-4-[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl]oxy-3-hydroxy-oxolan-2-yl]methoxy-hydroxy-phosphoryl] phosphono hydrogen phosphate | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4O1P
 
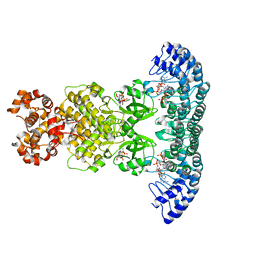 | | Crystal Structure of RNase L in complex with 2-5A and AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribonuclease L, ... | | Authors: | Huang, H, Zeqiraj, E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2013-12-16 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity.
Mol.Cell, 53, 2014
|
|
4DDI
 
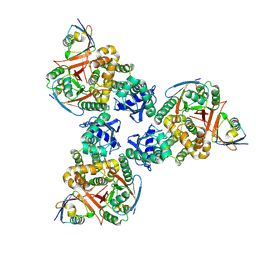 | | Crystal structure of human OTUB1/UbcH5b~Ub/Ub | | Descriptor: | Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D2, Ubiquitin thioesterase OTUB1 | | Authors: | Juang, Y.C, Sanches, M, Sicheri, F. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function.
Mol.Cell, 45, 2012
|
|
4DDG
 
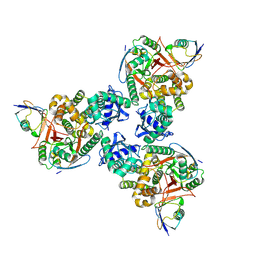 | | Crystal structure of human OTUB1/UbcH5b~Ub/Ub | | Descriptor: | Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 D2, Ubiquitin thioesterase OTUB1 | | Authors: | Juang, Y.C, Sanches, M, Sicheri, F. | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2987 Å) | | Cite: | OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function.
Mol.Cell, 45, 2012
|
|
7KJU
 
 | | Cgi121-tRNA complex | | Descriptor: | MAGNESIUM ION, RNA (75-MER) | | Authors: | Ceccarelli, D.F, Beenstock, J, Wan, L.C.K, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7KJT
 
 | | KEOPS tRNA modifying sub-complex of archaeal Cgi121 and tRNA | | Descriptor: | RNA (70-MER), Regulatory protein Cgi121 | | Authors: | Ceccarelli, D.F, Beenstock, J, Mao, D.Y.L, Sicheri, F. | | Deposit date: | 2020-10-26 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | A substrate binding model for the KEOPS tRNA modifying complex.
Nat Commun, 11, 2020
|
|
7LUO
 
 | | N-terminus of Skp2 bound to Cyclin A | | Descriptor: | S-phase kinase-associated protein 2,Cyclin-A2, Skp2 Motif 1 uncharacterized fragment 1, Skp2 Motif 1 uncharacterized fragment 2 | | Authors: | Kelso, S, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2021-02-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Bipartite binding of the N terminus of Skp2 to cyclin A.
Structure, 29, 2021
|
|
2K8Y
 
 | | Solution NMR Structure of Cgi121 from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MJ0187 | | Descriptor: | Uncharacterized protein MJ0187 | | Authors: | Rumpel, S, Fares, C, Neculai, D, Arrowsmith, C, Montelione, G.T, Sicheri, F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
3ENH
 
 | | Crystal structure of Cgi121/Bud32/Kae1 complex | | Descriptor: | HEXATANTALUM DODECABROMIDE, Putative O-sialoglycoprotein endopeptidase, Uncharacterized protein MJ0187 | | Authors: | Neculai, D. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Atomic Structure of the KEOPS Complex: An Ancient Protein Kinase-Containing Molecular Machine
Mol.Cell, 32, 2008
|
|
