6OCF
 
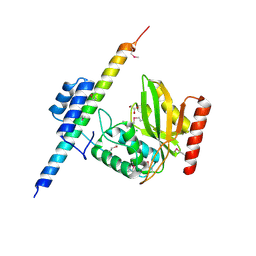 | | The crystal structure of VASH1-SVBP complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Small vasohibin-binding protein, ... | | Authors: | Li, F, Luo, X, Yu, H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural basis of tubulin detyrosination by vasohibins.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5WC2
 
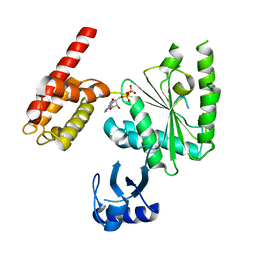 | | Crystal Structure of ADP-bound human TRIP13 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Jeong, B.-C, Luo, X. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
3GMH
 
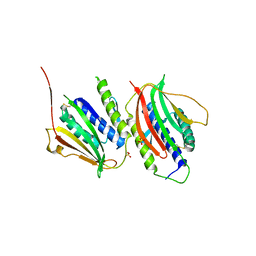 | | Crystal Structure of the Mad2 Dimer | | Descriptor: | Mitotic spindle assembly checkpoint protein MAD2A, SULFATE ION | | Authors: | Ozkan, E, Luo, X, Machius, M, Yu, H, Deisenhofer, J. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an intermediate conformer of the spindle checkpoint protein Mad2.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2IW5
 
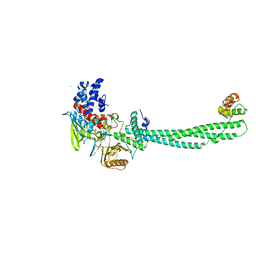 | | Structural Basis for CoREST-Dependent Demethylation of Nucleosomes by the Human LSD1 Histone Demethylase | | Descriptor: | AMMONIUM ION, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yang, M, Gocke, C.B, Luo, X, Borek, D, Tomchick, D.R, Machius, M, Otwinowski, Z, Yu, H. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Basis for Corest-Dependent Demethylation of Nucleosomes by the Human Lsd1 Histone Demethylase
Mol.Cell, 23, 2006
|
|
2QYF
 
 | |
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
2VFX
 
 | | Structure of the Symmetric Mad2 Dimer | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, ... | | Authors: | Yang, M, Li, B, Liu, C.-J, Tomchick, D.R, Machius, M, Rizo, J, Yu, H, Luo, X. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights Into MAD2 Regulation in the Spindle Checkpoint Revealed by the Crystal Structure of the Symmetric MAD2 Dimer.
Plos Biol., 6, 2008
|
|
5VZU
 
 | | Crystal structure of the Skp1-FBXO31-cyclin D1 complex | | Descriptor: | Cyclin D1, F-box only protein 31, PHOSPHATE ION, ... | | Authors: | Li, Y, Jin, K, Hao, B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the phosphorylation-independent recognition of cyclin D1 by the SCFFBXO31 ubiquitin ligase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6AFR
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with 5-((4-fluoro-1H-imidazol-1-yl)methyl)quinolin-8-ol | | Descriptor: | 5-[(4-fluoranylimidazol-1-yl)methyl]quinolin-8-ol, Bromodomain-containing protein 4 | | Authors: | Xing, J, Zhang, R.K, Zheng, M.Y, Luo, C, Jiang, X.R. | | Deposit date: | 2018-08-08 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Rational design of 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors
Eur J Med Chem, 163, 2018
|
|
5I2K
 
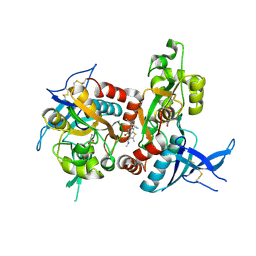 | | Structure of the human GluN1/GluN2A LBD in complex with 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 19) | | Descriptor: | 7-{[ethyl(4-fluorophenyl)amino]methyl}-N,2-dimethyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
1FYG
 
 | |
8UNH
 
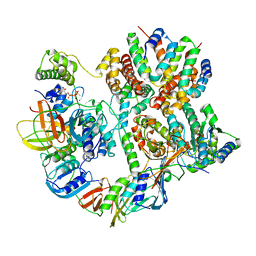 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-19 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8UNF
 
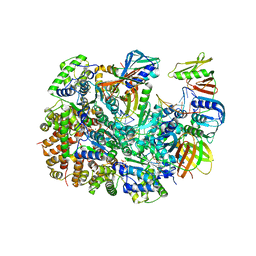 | | Cryo-EM structure of T4 Bacteriophage Clamp Loader with Sliding Clamp and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Sliding clamp, ... | | Authors: | Huang, Y, Marcus, K, Subramanian, S, Gee, L.C, Gorday, K, Ghaffari-Kashani, S, Luo, X, Zhang, L, O'Donnell, M, Subramanian, S, Kuriyan, J. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4QPM
 
 | | Structure of Bub1 kinase domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lin, Z.H, Jia, L.Y, Tomchick, D.R, Luo, X.L, Yu, H.T. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-22 | | Last modified: | 2014-12-24 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Substrate-Specific Activation of the Mitotic Kinase Bub1 through Intramolecular Autophosphorylation and Kinetochore Targeting.
Structure, 22, 2014
|
|
6WSL
 
 | | Cryo-EM structure of VASH1-SVBP bound to microtubules | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Small vasohibin-binding protein, ... | | Authors: | Li, F, Li, Y, Yu, H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of VASH1-SVBP bound to microtubules.
Elife, 9, 2020
|
|
7DMC
 
 | | Dipyridamole binds to the N-terminal domain of human Hsp90A | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, CHLORIDE ION, Heat shock protein HSP 90-alpha, ... | | Authors: | Shi, L, Zhou, C, Zhong, Y, Gao, J, Zhou, H, Zhang, N. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Dipyridamole interacts with the N-terminal domain of HSP90 and antagonizes the function of the chaperone in multiple cancer cell lines.
Biochem Pharmacol, 207, 2022
|
|
5I2N
 
 | | Structure of the human GluN1/GluN2A LBD in complex with N-ethyl-7-{[2-fluoro-3-(trifluoromethyl)phenyl]methyl}-2-methyl-5-oxo-5H-[1,3]thiazolo[3,2-a]pyrimidine-3-carboxamide (compound 29) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-02-09 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J.Med.Chem., 59, 2016
|
|
5KDT
 
 | | Structure of the human GluN1/GluN2A LBD in complex with GNE0723 | | Descriptor: | (1~{R},2~{R})-2-[7-[[5-chloranyl-3-(trifluoromethyl)pyrazol-1-yl]methyl]-5-oxidanylidene-2-(trifluoromethyl)-[1,3]thiazolo[3,2-a]pyrimidin-3-yl]cyclopropane-1-carbonitrile, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-08 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of GluN2A-Selective NMDA Receptor Positive Allosteric Modulators (PAMs): Tuning Deactivation Kinetics via Structure-Based Design.
J. Med. Chem., 59, 2016
|
|
5H25
 
 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5H24
 
 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
8HAY
 
 | | d4-bound btDPP4 | | Descriptor: | (1~{R})-1-[[4-[5-[[(1~{R})-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinolin-1-yl]methyl]-2-methoxy-phenoxy]phenyl]methyl]-6,7-dimethoxy-2-methyl-3,4-dihydro-1~{H}-isoquinoline, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-10-27 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
8HR7
 
 | | Structure of RdrA-RdrB complex | | Descriptor: | Adenosine deaminase, Archaeal ATPase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRB
 
 | | Structure of tetradecameric RdrA ring in RNA-loading state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Archaeal ATPase, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*GP*UP*CP*AP*UP*CP*GP*CP*UP*GP*GP*AP*C)-3') | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
8HRC
 
 | | Structure of dodecameric RdrB cage | | Descriptor: | Adenosine deaminase | | Authors: | Gao, Y. | | Deposit date: | 2022-12-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Molecular basis of RADAR anti-phage supramolecular assemblies.
Cell, 186, 2023
|
|
