4PJW
 
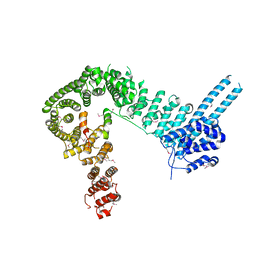 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1), with bound MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4PJU
 
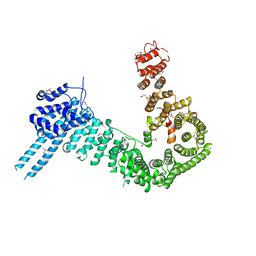 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) | | Descriptor: | Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-12 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3JA7
 
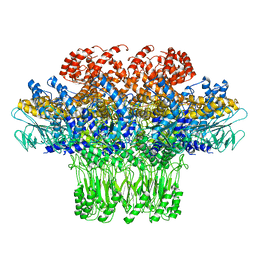 | | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution | | Descriptor: | Portal protein gp20 | | Authors: | Sun, L, Zhang, X, Gao, S, Rao, P.A, Padilla-Sanchez, V, Chen, Z, Sun, S, Xiang, Y, Subramaniam, S, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2015-04-21 | | Release date: | 2015-07-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution.
Nat Commun, 6, 2015
|
|
4RO0
 
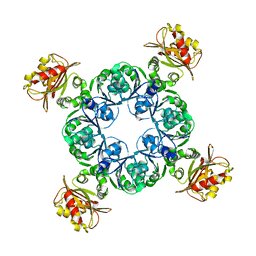 | | Crystal structure of MthK gating ring in a ligand-free form | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Dong, W, Guo, R, Chai, H, Chen, Z, Cui, H, Ren, Z, Li, Y, Ye, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The principle of cooperative gating in a calcium-gated K+ channel
To be Published
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6PXJ
 
 | | Crystal structure of human thrombin mutant I16T | | Descriptor: | GLYCEROL, MAGNESIUM ION, Thrombin heavy chain, ... | | Authors: | Stojanovski, B, Chen, Z, Koester, S.K, Pelc, L.A, Di Cera, E. | | Deposit date: | 2019-07-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the I16-D194 ionic interaction in the trypsin fold.
Sci Rep, 9, 2019
|
|
4PK7
 
 | | crystal structure of human Stromal Antigen 2 (SA2) in complex with Sister Chromatid Cohesion protein 1 (Scc1) with bound MES, native proteins | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cohesin subunit SA-2, Double-strand-break repair protein rad21 homolog | | Authors: | Hara, K, Chen, Z, Tomchick, D.R, Yu, H. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6JE4
 
 | | Crystal structure of Nme1Cas9-sgRNA-dsDNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | 1,2-ETHANEDIOL, AcrIIC3, CRISPR-associated endonuclease Cas9, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.069 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
4QKF
 
 | | Crystal structure of human ALKBH7 in complex with N-oxalylglycine and Mn(II) | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial, MANGANESE (II) ION, ... | | Authors: | Wang, G, Chen, Z. | | Deposit date: | 2014-06-06 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
J.Biol.Chem., 289, 2014
|
|
4QKB
 
 | | Crystal structure of seleno-methionine labelled human ALKBH7 in complex with alpha-ketoglutarate and Mn(II) | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 7, mitochondrial, ... | | Authors: | Wang, G, He, Q, Chen, Z. | | Deposit date: | 2014-06-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The atomic resolution structure of human AlkB homolog 7 (ALKBH7), a key protein for programmed necrosis and fat metabolism
J.Biol.Chem., 289, 2014
|
|
4QKD
 
 | |
2H47
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
6A58
 
 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
1Z71
 
 | | thrombin and P2 pyridine N-oxide inhibitor complex structure | | Descriptor: | Hirudin IIIB', L17, thrombin | | Authors: | Nantermet, P.G, Burgey, C.S, Robinson, K.A, Pellicore, J.M, Newton, C.L, Deng, J.Z, Lyle, T.A, Selnick, H.G, Lewis, S.D, Lucas, B.J, Krueger, J.A, Miller-Stein, C, White, R.B, Wong, B, McMasters, D.R, Wallace, A.A, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P(2) pyridine N-oxide thrombin inhibitors: a novel peptidomimetic scaffold
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
3BEF
 
 | | Crystal structure of thrombin bound to the extracellular fragment of PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Prothrombin | | Authors: | Gandhi, P.S, Bah, A, Chen, Z, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-11-17 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural identification of the pathway of long-range communication in an allosteric enzyme.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4RKJ
 
 | | Crystal structure of thrombin mutant S195T (free form) | | Descriptor: | GLYCEROL, POTASSIUM ION, Thrombin heavy chain, ... | | Authors: | Pelc, A.L, Chen, Z, Gohara, D.W, Vogt, A.D, Pozzi, N, Di Cera, E. | | Deposit date: | 2014-10-13 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Why ser and not thr brokers catalysis in the trypsin fold.
Biochemistry, 54, 2015
|
|
4NRM
 
 | |
4RN6
 
 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | Descriptor: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | Deposit date: | 2014-10-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
