3UJ0
 
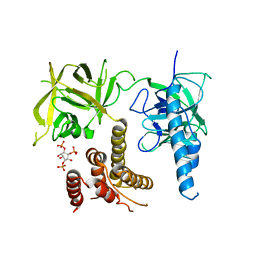 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor with ligand bound form. | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | 登録日 | 2011-11-07 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
2BBN
 
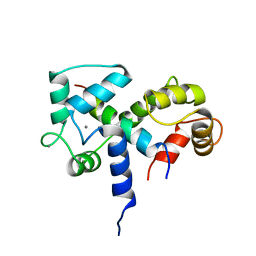 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
2BBM
 
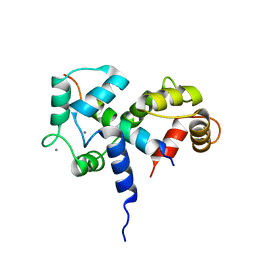 | | SOLUTION STRUCTURE OF A CALMODULIN-TARGET PEPTIDE COMPLEX BY MULTIDIMENSIONAL NMR | | 分子名称: | CALCIUM ION, CALMODULIN, MYOSIN LIGHT CHAIN KINASE | | 著者 | Clore, G.M, Bax, A, Ikura, M, Gronenborn, A.M. | | 登録日 | 1992-07-16 | | 公開日 | 1994-01-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a calmodulin-target peptide complex by multidimensional NMR.
Science, 256, 1992
|
|
3UJ4
 
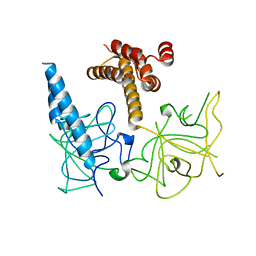 | | Crystal structure of the apo-inositol 1,4,5-trisphosphate receptor | | 分子名称: | Inositol 1,4,5-trisphosphate receptor type 1, SULFATE ION | | 著者 | Ikura, M, Seo, M.D, Ishiyama, N, Stathopulos, P. | | 登録日 | 2011-11-07 | | 公開日 | 2012-02-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and functional conservation of key domains in InsP3 and ryanodine receptors.
Nature, 483, 2012
|
|
8VM2
 
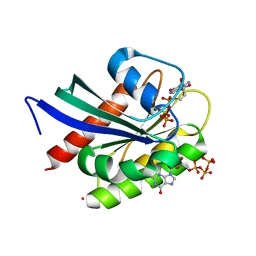 | | Crystal Structure of NRAS Q61K bound to GTP | | 分子名称: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Gebregiworgis, T, Chan, J.Y.L, Kuntz, D.A, Prive, G.G, Marshall, C.B, Ikura, M. | | 登録日 | 2024-01-12 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Eur J Cell Biol, 103, 2024
|
|
1N4K
 
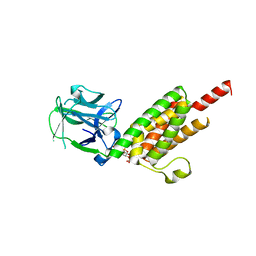 | | Crystal structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with IP3 | | 分子名称: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | 著者 | Bosanac, I, Alattia, J.R, Mal, T.K, Chan, J, Talarico, S, Tong, F.K, Tong, K.I, Yoshikawa, F, Furuichi, T, Iwai, M, Michikawa, T, Mikoshiba, K, Ikura, M. | | 登録日 | 2002-10-31 | | 公開日 | 2002-12-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the inositol 1,4,5-trisphosphate receptor
binding core in complex with its ligand.
Nature, 420, 2002
|
|
7SCW
 
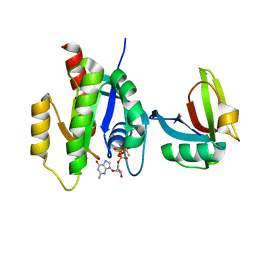 | | KRAS full length wild-type in complex with RGL1 Ras association domain | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | 著者 | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | 登録日 | 2021-09-29 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
7SCX
 
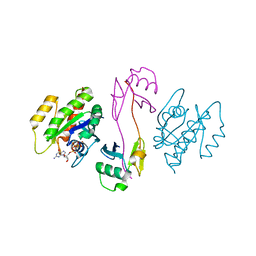 | | KRAS full-length G12V in complex with RGL1 Ras association domain | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | 著者 | Eves, B.J, Kuntz, D.A, Ikura, M, Marshall, C.B. | | 登録日 | 2021-09-29 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Structures of RGL1 RAS-Association Domain in Complex with KRAS and the Oncogenic G12V Mutant.
J.Mol.Biol., 434, 2022
|
|
3L6Y
 
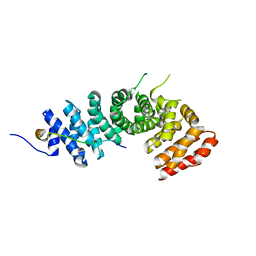 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
1MYW
 
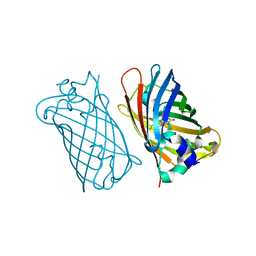 | | CRYSTAL STRUCTURE OF A YELLOW FLUORESCENT PROTEIN WITH IMPROVED MATURATION AND REDUCED ENVIRONMENTAL SENSITIVITY | | 分子名称: | Green fluorescent protein | | 著者 | Rekas, A, Alattia, J.R, Nagai, T, Miyawaki, A, Ikura, M. | | 登録日 | 2002-10-04 | | 公開日 | 2003-01-14 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Venus, a Yellow Fluorescent
Protein with Improved Maturation and
Reduced Environmental Sensitivity
J.Biol.Chem., 277, 2002
|
|
6OS4
 
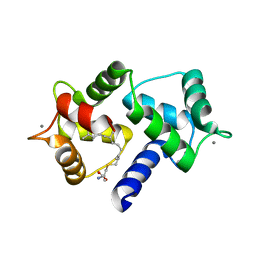 | | Calmodulin in complex with farnesyl cysteine methyl ester | | 分子名称: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | 著者 | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | 登録日 | 2019-05-01 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
5WI4
 
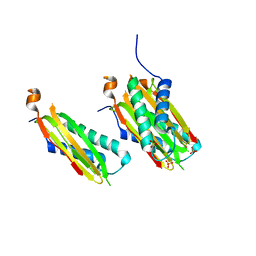 | | CRYSTAL STRUCTURE OF DYNLT1/TCTEX-1 IN COMPLEX WITH ARHGEF2 | | 分子名称: | Dynein light chain Tctex-type 1,Rho guanine nucleotide exchange factor 2, SULFATE ION | | 著者 | Balan, M, Ishiyama, N, Marshall, C.B, Ikura, M. | | 登録日 | 2017-07-18 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | MARK3-mediated phosphorylation of ARHGEF2 couples microtubules to the actin cytoskeleton to establish cell polarity.
Sci Signal, 10, 2017
|
|
4B0A
 
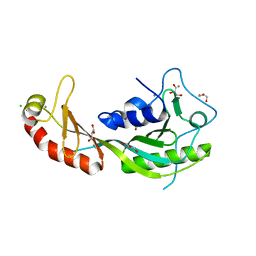 | | The high-resolution structure of yTBP-yTAF1 identifies conserved and competing interaction surfaces in transcriptional activation | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Anandapadamanaban, M, Andresen, C, Siponen, M, Kokubo, T, Ikura, M, Moche, M, Sunnerhagen, M. | | 登録日 | 2012-06-29 | | 公開日 | 2013-07-03 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | High-Resolution Structure of TBP with Taf1 Reveals Anchoring Patterns in Transcriptional Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
6DUW
 
 | |
6DV1
 
 | |
6DUY
 
 | |
6W4E
 
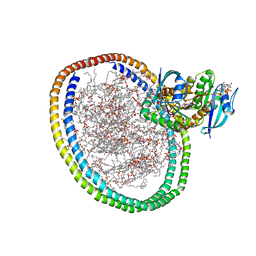 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | 著者 | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | 登録日 | 2020-03-10 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | 登録日 | 2020-03-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7RSC
 
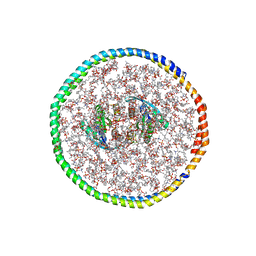 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSE
 
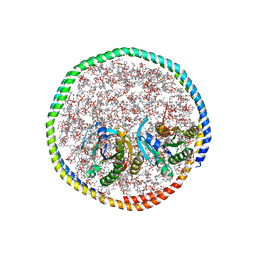 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
3L6X
 
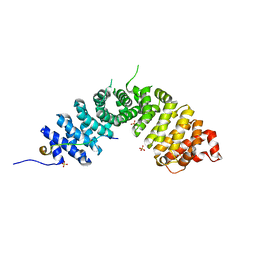 | | Crystal structure of p120 catenin in complex with E-cadherin | | 分子名称: | Catenin delta-1, E-cadherin, SULFATE ION | | 著者 | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | 登録日 | 2009-12-27 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
6PW7
 
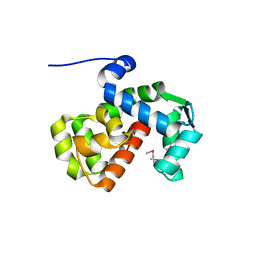 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | 分子名称: | CALCIUM ION, Stromal interaction molecule 1 | | 著者 | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | 登録日 | 2019-07-22 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-02-12 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
1IQ5
 
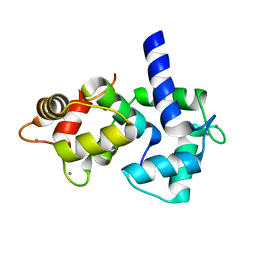 | | Calmodulin/nematode CA2+/Calmodulin dependent kinase kinase fragment | | 分子名称: | CA2+/CALMODULIN DEPENDENT KINASE KINASE, CALCIUM ION, CALMODULIN | | 著者 | Kurokawa, H, Osawa, M, Kurihara, H, Katayama, N, Tokumitsu, H, Swindells, M.B, Kainosho, M, Ikura, M. | | 登録日 | 2001-06-14 | | 公開日 | 2001-09-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Target-induced conformational adaptation of calmodulin revealed by the crystal structure of a complex with nematode Ca(2+)/calmodulin-dependent kinase kinase peptide
J.Mol.Biol., 312, 2001
|
|
6AMB
 
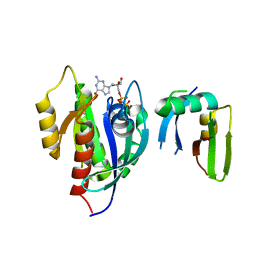 | | Crystal Structure of the Afadin RA1 domain in complex with HRAS | | 分子名称: | Afadin, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Smith, M.J, Ishiyama, N, Ikura, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-11-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Evolution of AF6-RAS association and its implications in mixed-lineage leukemia.
Nat Commun, 8, 2017
|
|
3HSM
 
 | | Crystal structure of distal N-terminal beta-trefoil domain of Ryanodine Receptor type 1 | | 分子名称: | Ryanodine receptor 1 | | 著者 | Amador, F.J, Liu, S, Ishiyama, N, Plevin, M.J, Wilson, A, MacLennan, D.H, Ikura, M. | | 登録日 | 2009-06-10 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of type I ryanodine receptor amino-terminal beta-trefoil domain reveals a disease-associated mutation "hot spot" loop
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
