3LJW
 
 | | Crystal Structure of the Second Bromodomain of Human Polybromo | | Descriptor: | ACETATE ION, Protein polybromo-1, SODIUM ION | | Authors: | Charlop-Powers, Z, Zhou, M.M, Zeng, L, Zhang, Q. | | Deposit date: | 2010-01-26 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2.
Cell Res., 20, 2010
|
|
6DNE
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS660 | | Descriptor: | Bromodomain-containing protein 4, N,N'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DJC
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2RNW
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNX
 
 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RNY
 
 | | Complex Structures of CBP Bromodomain with H4 ack20 Peptide | | Descriptor: | CREB-binding protein, Histone H4 | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
1SZV
 
 | | Structure of the Adaptor Protein p14 reveals a Profilin-like Fold with Novel Function | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein | | Authors: | Qian, C, Zhang, Q, Wang, X, Zeng, L, Farooq, A, Zhou, M.M. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Adaptor Protein p14 Reveals a Profilin-like Fold with Distinct Function
J.Mol.Biol., 347, 2005
|
|
1WUG
 
 | | complex structure of PCAF bromodomain with small chemical ligand NP1 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-4-METHYL-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-07 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
1WUM
 
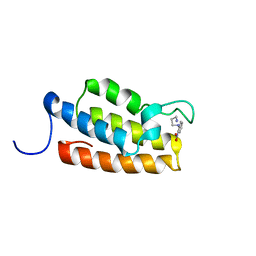 | | Complex structure of PCAF bromodomain with small chemical ligand NP2 | | Descriptor: | Histone acetyltransferase PCAF, N-(3-AMINOPROPYL)-2-NITROBENZENAMINE | | Authors: | Zeng, L, Li, J, Muller, M, Yan, S, Mujtaba, S, Pan, C, Wang, Z, Zhou, M.M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-08-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Selective small molecules blocking HIV-1 Tat and coactivator PCAF association
J.Am.Chem.Soc., 127, 2005
|
|
6DUV
 
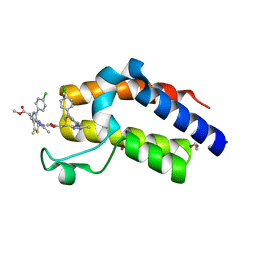 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
3KMT
 
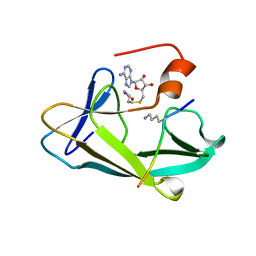 | | Crystal structure of vSET/SAH/H3 ternary complex | | Descriptor: | A612L protein, Histone H3, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Wei, H, Zhou, M.M. | | Deposit date: | 2009-11-11 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Dimerization of a viral SET protein endows its function.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KMJ
 
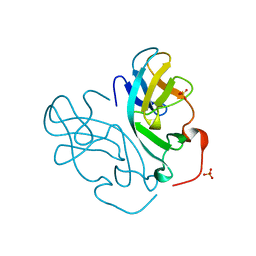 | | Crystal structure of vSET under condition B | | Descriptor: | A612L protein, SULFATE ION | | Authors: | Wei, H, Zhou, M.M. | | Deposit date: | 2009-11-10 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dimerization of a viral SET protein endows its function.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KMA
 
 | | Crystal Structure of vSET under Condition A | | Descriptor: | A612L protein, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wei, H, Zhou, M.M. | | Deposit date: | 2009-11-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerization of a viral SET protein endows its function.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2D82
 
 | | Target Structure-Based Discovery of Small Molecules that Block Human p53 and CREB Binding Protein (CBP) Association | | Descriptor: | 9-ACETYL-2,3,4,9-TETRAHYDRO-1H-CARBAZOL-1-ONE, CREB-binding protein | | Authors: | Sachchidanand, Resnick-Silverman, L, Yan, S, Mujtaba, S, Liu, W.J, Zeng, L, Manfredi, J.J, Zhou, M.M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Target structure-based discovery of small molecules that block human p53 and CREB binding protein association
Chem.Biol., 13, 2006
|
|
2G46
 
 | |
5EJW
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS351 | | Descriptor: | (1~{R})-2-[2-azanylidene-3-[(2-methylphenyl)methyl]benzimidazol-1-yl]-1-(3,4-dichlorophenyl)ethanol, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Discovery of Selective Antagonists for the Chromodomain of Polycomb Repressive Protein CBX7.
Acs Med.Chem.Lett., 7, 2016
|
|
4X3S
 
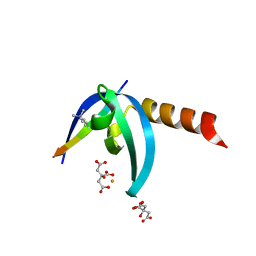 | | Crystal structure of chromobox homology 7 (CBX7) with SETDB1-1170me3 Peptide | | Descriptor: | CITRIC ACID, Chromobox protein homolog 7, FE (III) ION, ... | | Authors: | Ren, C, Plotnikov, A.N, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3T
 
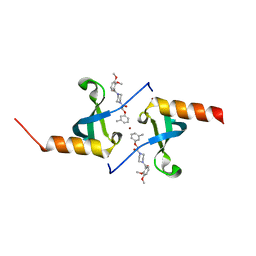 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS37452 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2,3-dimethoxybenzoyl)piperazin-1-yl]-2-(3-methylphenoxy)ethanone, Chromobox protein homolog 7, ... | | Authors: | Ren, C, Jakoncic, J, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
4X3K
 
 | |
4X3U
 
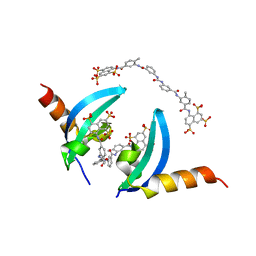 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with Suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2014-12-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Small-Molecule Modulators of Methyl-Lysine Binding for the CBX7 Chromodomain.
Chem.Biol., 22, 2015
|
|
1SHC
 
 | | SHC PTB DOMAIN COMPLEXED WITH A TRKA RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | SHC, TRKA RECEPTOR PHOSPHOPEPTIDE | | Authors: | Zhou, M.-M, Ravichandran, K.S, Olejniczak, E.T, Petros, A.M, Meadows, R.P, Sattler, M, Harlan, J.E, Wade, W.S, Burakoff, S.J, Fesik, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the phosphotyrosine binding domain of Shc.
Nature, 378, 1995
|
|
1TCE
 
 | | SOLUTION NMR STRUCTURE OF THE SHC SH2 DOMAIN COMPLEXED WITH A TYROSINE-PHOSPHORYLATED PEPTIDE FROM THE T-CELL RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PHOSPHOPEPTIDE OF THE ZETA CHAIN OF T CELL RECEPTOR, SHC | | Authors: | Zhou, M.-M, Meadows, R.P, Logan, T.M, Yoon, H.S, Wade, W.R, Ravichandran, K.S, Burakoff, S.J, Feisk, S.W. | | Deposit date: | 1996-03-27 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Shc SH2 domain complexed with a tyrosine-phosphorylated peptide from the T-cell receptor.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1IRS
 
 | | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IL-4 RECEPTOR PHOSPHOPEPTIDE, IRS-1 | | Authors: | Zhou, M.-M, Huang, B, Olejniczak, E.T, Meadows, R.P, Shuker, S.B, Miyazaki, M, Trub, T, Shoelson, S.E, Feisk, S.W. | | Deposit date: | 1996-03-22 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain.
Nat.Struct.Biol., 3, 1996
|
|
5ULA
 
 | |
5U5S
 
 | |
