3B2Z
 
 | | Crystal Structure of ADAMTS4 (apo form) | | Descriptor: | ADAMTS-4, CALCIUM ION, ZINC ION | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2ID5
 
 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
2RJQ
 
 | | Crystal structure of ADAMTS5 with inhibitor bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, ADAMTS-5, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
2RJP
 
 | | Crystal structure of ADAMTS4 with inhibitor bound | | Descriptor: | ADAMTS-4, CALCIUM ION, N-({4'-[(4-isobutyrylphenoxy)methyl]biphenyl-4-yl}sulfonyl)-D-valine, ... | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-15 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
7KQK
 
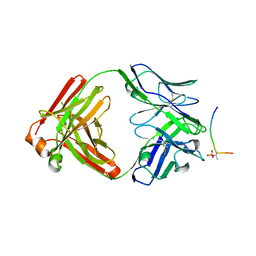 | |
1F46
 
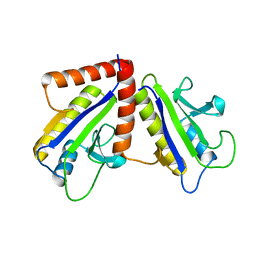 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1F47
 
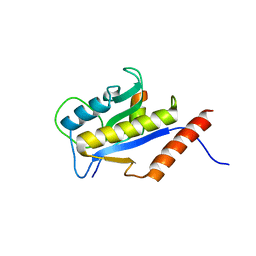 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | CELL DIVISION PROTEIN FTSZ, CELL DIVISION PROTEIN ZIPA | | Authors: | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | Deposit date: | 2000-06-07 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
5FCS
 
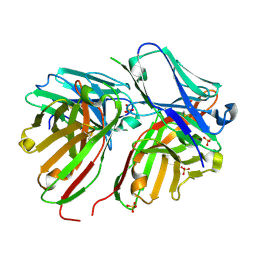 | | Diabody | | Descriptor: | Diabody, SULFATE ION | | Authors: | Mosyak, L, Root, A. | | Deposit date: | 2015-12-15 | | Release date: | 2016-12-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of PF-06671008, a Highly Potent Anti-P-cadherin/Anti-CD3 Bispecific DART Molecule with Extended Half-Life for the Treatment of Cancer.
Antibodies, 5, 2016
|
|
8GHP
 
 | | GUCY2C-ECD bound to anti-GUCY2C-scFv antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Guanylyl cyclase C, anti-GUCY2C-scFv antibody heavy chain, ... | | Authors: | Mosyak, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Molecular insights into recognition of GUCY2C by T-cell engaging bispecific antibody anti-GUCY2CxCD3.
Sci Rep, 13, 2023
|
|
8GHO
 
 | | GUCY2C-peptide bound to anti-GUCY2C-scFv antibody | | Descriptor: | Guanylyl cyclase C peptide, anti-GUCY2C-scFv antibody heavy chain, anti-GUCY2C-scFv antibody light chain | | Authors: | Mosyak, L. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into recognition of GUCY2C by T-cell engaging bispecific antibody anti-GUCY2CxCD3.
Sci Rep, 13, 2023
|
|
1Y2F
 
 | | Crystal Structure of ZipA with an inhibitor | | Descriptor: | 4-{2-[4-(2-AMINOETHYL)PIPERAZIN-1-YL]PYRIDIN-4-YL}-N-(3-CHLORO-4-METHYLPHENYL)PYRIMIDIN-2-AMINE, Cell division protein zipA | | Authors: | Mosyak, L, Rush, T.S. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction
J.Med.Chem., 48, 2005
|
|
1Y2G
 
 | | Crystal STructure of ZipA in complex with an inhibitor | | Descriptor: | Cell division protein zipA, N-METHYL-N-[3-(6-PHENYL[1,2,4]TRIAZOLO[4,3-B]PYRIDAZIN-3-YL)PHENYL]ACETAMIDE | | Authors: | Mosyak, L, Rush, T.S. | | Deposit date: | 2004-11-22 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A shape-based 3-D scaffold hopping method and its application to a bacterial protein-protein interaction
J.Med.Chem., 48, 2005
|
|
1PYS
 
 | | PHENYLALANYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS | | Descriptor: | MAGNESIUM ION, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Safro, M, Mosyak, L, Goldgur, Y, Reshetnikova, L, Delarue, M. | | Deposit date: | 1996-11-14 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of phenylalanyl-tRNA synthetase from Thermus thermophilus.
Nat.Struct.Biol., 2, 1995
|
|
7KQ7
 
 | | Crystal structure of IL21R in complex with an antibody Fab fragment | | Descriptor: | Antibody heavy chain, Antibody light chain, Interleukin-21 receptor | | Authors: | Mosyak, L, Svenson, K. | | Deposit date: | 2020-11-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Combining random mutagenesis, structure-guided design and next-generation sequencing to mitigate polyreactivity of an anti-IL-21R antibody.
Mabs, 13, 2021
|
|
1HDM
 
 | | HISTOCOMPATIBILITY ANTIGEN HLA-DM | | Descriptor: | PROTEIN (CLASS II HISTOCOMPATIBILITY ANTIGEN, M ALPHA CHAIN), M BETA CHAIN) | | Authors: | Wiley, D.C, Mosyak, L. | | Deposit date: | 1998-09-09 | | Release date: | 1999-09-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation.
Immunity, 9, 1998
|
|
6WIV
 
 | | Structure of human GABA(B) receptor in an inactive state | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-{[(9Z)-octadec-9-enoyl]oxy}propyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Park, J, Fu, Z, Frangaj, A, Liu, J, Mosyak, L, Shen, T, Slavkovich, V.N, Ray, K.M, Taura, J, Cao, B, Geng, Y, Zuo, H, Kou, Y, Grassucci, R, Chen, S, Liu, Z, Lin, X, Williams, J.P, Rice, W.J, Eng, E.T, Huang, R.K, Soni, R.K, Kloss, B, Yu, Z, Javitch, J.A, Hendrickson, W.A, Slesinger, P.A, Quick, M, Graziano, J, Yu, H, Fiehn, O, Clarke, O.B, Frank, J, Fan, Q.R. | | Deposit date: | 2020-04-10 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of human GABABreceptor in an inactive state.
Nature, 584, 2020
|
|
6OCP
 
 | | Crystal structure of a human GABAB receptor peptide bound to KCTD16 T1 | | Descriptor: | BTB/POZ domain-containing protein KCTD16, Gamma-aminobutyric acid type B receptor subunit 2 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3KXZ
 
 | | The complex crystal structure of LCK with a probe molecule w259 | | Descriptor: | 3-[7-[(3-hydroxyphenyl)amino]pyrazolo[1,5-a]pyrimidin-2-yl]-N-(1-hydroxy-2,2,6,6-tetramethyl-piperidin-4-yl)benzamide, CALCIUM ION, Proto-oncogene tyrosine-protein kinase LCK, ... | | Authors: | Xu, Z.B, Moy, F.J, Kelleher, K, Mosyak, L, Protein Structure Factory (PSF) | | Deposit date: | 2009-12-04 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Novel synthesis and structural characterization of a high-affinity paramagnetic kinase probe for the identification of non-ATP site binders by nuclear magnetic resonance.
J.Med.Chem., 53, 2010
|
|
1SQ0
 
 | | Crystal Structure of the Complex of the Wild-type Von Willebrand Factor A1 domain and Glycoprotein Ib alpha at 2.6 Angstrom Resolution | | Descriptor: | Platelet glycoprotein Ib alpha chain (Glycoprotein Ibalpha) (GP-Ib alpha) (GPIbA) (GPIb-alpha) (CD42B-alpha) (CD42B) [Contains: Glycocalicin], Von Willebrand factor (vWF) [Contains: Von Willebrand antigen II] | | Authors: | Dumas, J.J, Kumar, R, McDonagh, T, Sullivan, F, Stahl, M.L, Somers, W.S, Mosyak, L. | | Deposit date: | 2004-03-17 | | Release date: | 2004-04-13 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations
J.Biol.Chem., 279, 2004
|
|
5HDQ
 
 | | MntC co-structure with mAB 305-78-7 | | Descriptor: | ABC transporter substrate-binding protein, FE (III) ION, Fab Heavy Chain, ... | | Authors: | Parris, K, Mosyak, L. | | Deposit date: | 2016-01-05 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High Resolution Mapping of Bactericidal Monoclonal Antibody Binding Epitopes on Staphylococcus aureus Antigen MntC.
Plos Pathog., 12, 2016
|
|
1I8L
 
 | | HUMAN B7-1/CTLA-4 CO-STIMULATORY COMPLEX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC T-LYMPHOCYTE PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD80, ... | | Authors: | Stamper, C.C, Somers, W.S, Mosyak, L. | | Deposit date: | 2001-03-14 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses.
Nature, 410, 2001
|
|
5K5S
 
 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
6OCR
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5K5T
 
 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
