8GY0
 
 | | Agrocybe pediades linalool sunthase (Ap.LS) | | Descriptor: | Terpene synthase | | Authors: | Rehka, T, Sharma, D, Lin, F, Lim, C, Choong, Y.K, Chacko, J, Zhang, C. | | Deposit date: | 2022-09-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.988 Å) | | Cite: | Structural Understanding of Fungal Terpene Synthases for the Formation of Linear or Cyclic Terpene Products.
Acs Catalysis, 13, 2023
|
|
8IKL
 
 | | Cryo-EM structure of the CD97-G13 complex | | Descriptor: | Adhesion G protein-coupled receptor E5, Guanine nucleotide-binding protein G(13) subunit alpha isoforms short, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Zhao, R, Dong, Y, Gao, M, Chen, L, Zhang, C, Xiao, P. | | Deposit date: | 2023-02-28 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Conformational transitions and activation of the adhesion receptor CD97.
Mol.Cell, 84, 2024
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
7VWP
 
 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
6KXH
 
 | | Alp1U_Y247F mutant in complex with Fluostatin C | | Descriptor: | D-MALATE, Fluostatin C, Putative hydrolase, ... | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78039551 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | Descriptor: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CX4
 
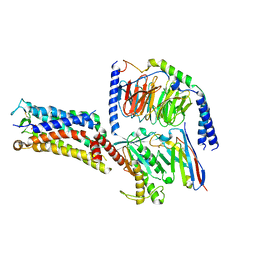 | | Cryo-EM structure of the Evatanepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-~{tert}-butylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX3
 
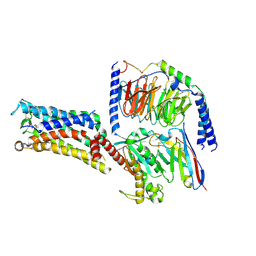 | | Cryo-EM structure of the Taprenepag-bound EP2-Gs complex | | Descriptor: | 2-[3-[[(4-pyrazol-1-ylphenyl)methyl-pyridin-3-ylsulfonyl-amino]methyl]phenoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7CX2
 
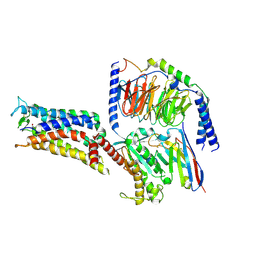 | | Cryo-EM structure of the PGE2-bound EP2-Gs complex | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qu, C, Mao, C, Xiao, P, Shen, Q, Zhong, Y, Yang, F, Shen, D, Tao, X, Zhang, H, Yan, X, Zhao, R, He, J, Guan, Y, Zhang, C, Hou, G, Zhang, P, Yu, X, Guan, Y, Sun, J, Zhang, Y. | | Deposit date: | 2020-09-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ligand recognition, unconventional activation, and G protein coupling of the prostaglandin E 2 receptor EP2 subtype.
Sci Adv, 7, 2021
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
5L6O
 
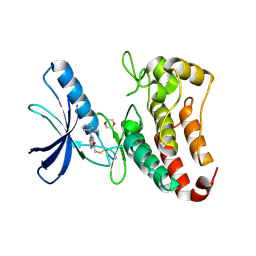 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 3) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 1-(4-phenylazanylquinazolin-7-yl)ethanone, Ephrin type-B receptor 3 | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
3FCU
 
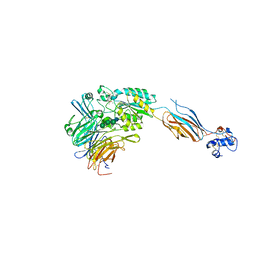 | | Structure of headpiece of integrin aIIBb3 in open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3FCS
 
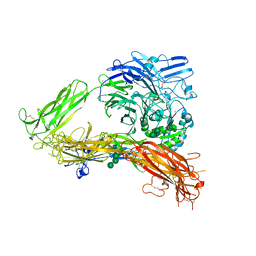 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3FBV
 
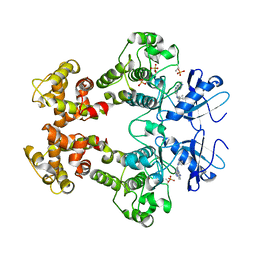 | | Crystal structure of the oligomer formed by the kinase-ribonuclease domain of Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A.V, Egea, P.F, Korostelev, A.A, Finer-Moore, J, Zhang, C, Shokat, K.M, Stroud, R.M, Walter, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The unfolded protein response signals through high-order assembly of Ire1.
Nature, 457, 2009
|
|
5MJA
 
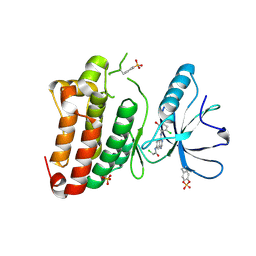 | | Kinase domain of human EphB1 bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
5L6P
 
 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 6) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ephrin type-B receptor 3, ~{N}-(4-phenylazanylquinazolin-7-yl)ethanamide | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
5MJB
 
 | | Kinase domain of human EphB1, G703C mutant, covalently bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
2G15
 
 | | Structural Characterization of autoinhibited c-Met kinase | | Descriptor: | activated met oncogene | | Authors: | Wang, W, Marimuthu, A, Tsai, J, Kumar, A, Krupka, H.I, Zhang, C, Powell, B, Suzuki, Y, Nguyen, H, Tabrizizad, M, Luu, C, West, B.L. | | Deposit date: | 2006-02-13 | | Release date: | 2006-03-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural characterization of autoinhibited c-Met kinase produced by coexpression in bacteria with phosphatase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
7V1U
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZJ12 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-2-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
7V2J
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 33 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-(3-ethyl-6-methoxy-1,2-benzoxazol-5-yl)-4-methoxy-benzenesulfonamide | | Authors: | Zhang, M, Wang, C, Zhang, C, Zhang, Y, Xu, Y. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Design, synthesis and pharmacological characterization of N-(3-ethylbenzo[d]isoxazol-5-yl) sulfonamide derivatives as BRD4 inhibitors against acute myeloid leukemia.
Acta Pharmacol.Sin., 43, 2022
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | Descriptor: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | Authors: | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | Deposit date: | 2020-06-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
