8JCZ
 
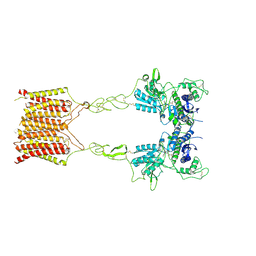 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode III) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCV
 
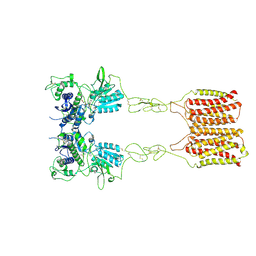 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCU
 
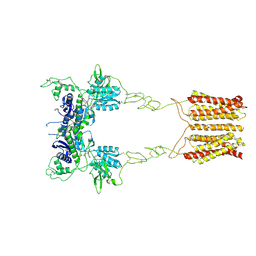 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD5
 
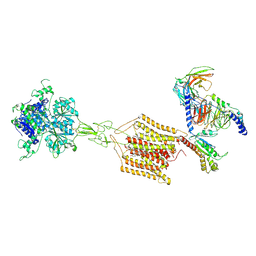 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD0
 
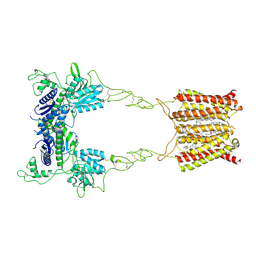 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of NAM563 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(1-methylpyrazol-4-yl)-7-[[(2~{S})-2-(trifluoromethyl)morpholin-4-yl]methyl]quinoline-2-carboxamide, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCX
 
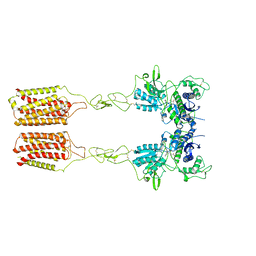 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495 and NAM563 (dimerization mode II) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JCY
 
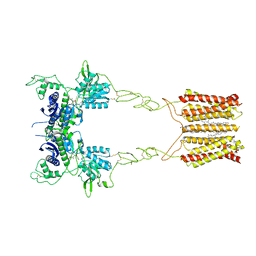 | | Cryo-EM structure of mGlu2-mGlu3 heterodimer in presence of LY341495, NAM563, and LY2389575 (dimerization mode I) | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD3
 
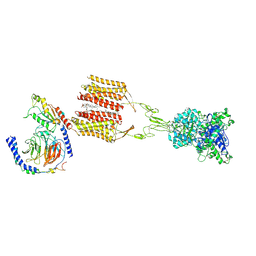 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD4
 
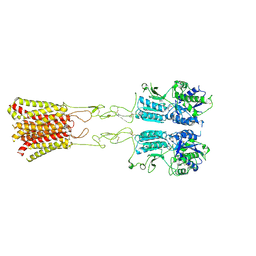 | | Cryo-EM structure of G protein-free mGlu2-mGlu4 heterodimer in Acc state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, Metabotropic glutamate receptor 2,Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
7X9B
 
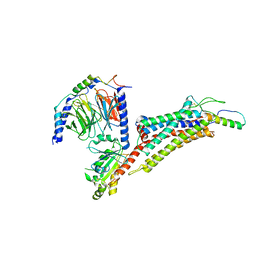 | | Cryo-EM structure of neuropeptide Y Y2 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7X9A
 
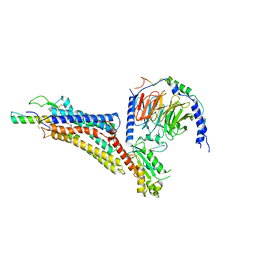 | | Cryo-EM structure of neuropeptide Y Y1 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
7X9C
 
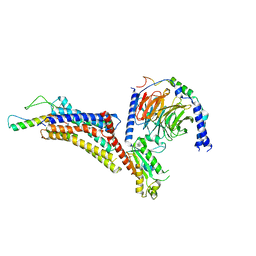 | | Cryo-EM structure of neuropeptide Y Y4 receptor in complex with PP and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Tang, T, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Receptor-specific recognition of NPY peptides revealed by structures of NPY receptors.
Sci Adv, 8, 2022
|
|
5YQZ
 
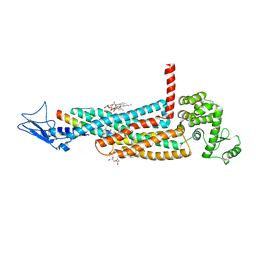 | | Structure of the glucagon receptor in complex with a glucagon analogue | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon analogue, ... | | Authors: | Zhang, H, Qiao, A, Yang, L, VAN EPS, N, Frederiksen, K, Yang, D, Dai, A, Cai, X, Zhang, H, Yi, C, Can, C, He, L, Yang, H, Lau, J, Ernst, O, Hanson, M, Stevens, R, Wang, M, Seedtz-Runge, S, Jiang, H, Zhao, Q, Wu, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the glucagon receptor in complex with a glucagon analogue.
Nature, 553, 2018
|
|
5Z0V
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-21 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
5ZBH
 
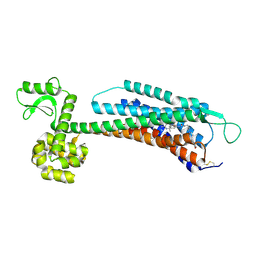 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5ZBQ
 
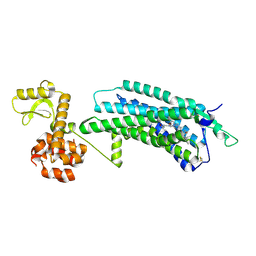 | | The Crystal Structure of human neuropeptide Y Y1 receptor with UR-MK299 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme, N~2~-(diphenylacetyl)-N-[(4-hydroxyphenyl)methyl]-N~5~-(N'-{[2-(propanoylamino)ethyl]carbamoyl}carbamimidoyl)-D-ornithinamide | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
5XEZ
 
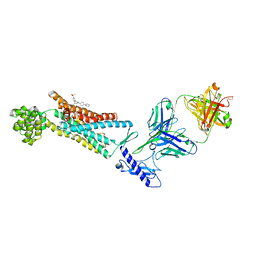 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-cyclohexylphenyl){[3-(methylsulfonyl)phenyl]carbamoyl}amino]methyl}-N-(1H-tetrazol-5-yl)benzamide, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
7F1R
 
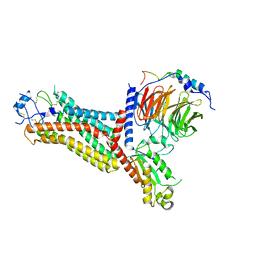 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with RANTES and Gi | | Descriptor: | C-C motif chemokine 5,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1S
 
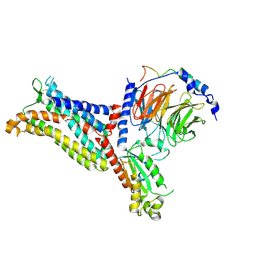 | | Cryo-EM structure of the apo chemokine receptor CCR5 in complex with Gi | | Descriptor: | C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1T
 
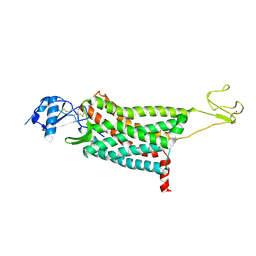 | | Crystal structure of the human chemokine receptor CCR5 in complex with MIP-1a | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ZINC ION | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7F1Q
 
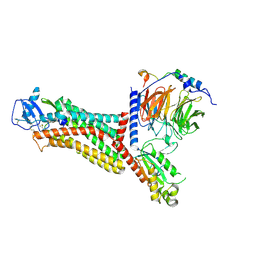 | | Cryo-EM structure of the chemokine receptor CCR5 in complex with MIP-1a and Gi | | Descriptor: | C-C motif chemokine 3,C-C chemokine receptor type 5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, H, Chen, K, Tan, Q, Han, S, Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for chemokine recognition and receptor activation of chemokine receptor CCR5.
Nat Commun, 12, 2021
|
|
7WVY
 
 | | Cryo-EM structure of the human formyl peptide receptor 2 in complex with Abeta42 and Gi2 | | Descriptor: | Amyloid-beta A4 protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhu, Y, Lin, X, Zong, X, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2022-02-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of FPR2 in recognition of A beta 42 and neuroprotection by humanin.
Nat Commun, 13, 2022
|
|
