8R0A
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ss complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QPE
 
 | | Cryo-EM Structure of Pre-B-like Complex (core part) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, INOSITOL HEXAKISPHOSPHATE, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-01 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8QZS
 
 | | Cryo-EM structure of the cross-exon B-like complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Microfibrillar-associated protein 1, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
6BI6
 
 | | Solution NMR structure of uncharacterized protein YejG | | Descriptor: | Uncharacterized protein YejG | | Authors: | Mohanty, B, Finn, T.J, Macindoe, I, Zhong, J, Patrick, W.M, Mackay, J.P. | | Deposit date: | 2017-11-01 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The uncharacterized bacterial protein YejG has the same architecture as domain III of elongation factor G.
Proteins, 87, 2019
|
|
6AEI
 
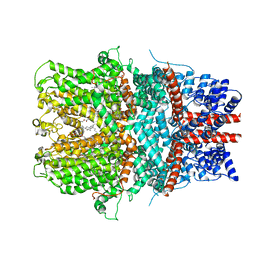 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
4XWY
 
 | | Crystal structure of human sepiapterin reductase in complex with an N-acetylserotinin analogue | | Descriptor: | N-[2-(5-hydroxy-2-methyl-1H-indol-3-yl)ethyl]-2-methoxyacetamide, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Johnsson, K, Hovius, R, Gorszka, K.I, Pojer, F. | | Deposit date: | 2015-01-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Reduction of Neuropathic and Inflammatory Pain through Inhibition of the Tetrahydrobiopterin Pathway.
Neuron, 86, 2015
|
|
1LYP
 
 | |
8XR4
 
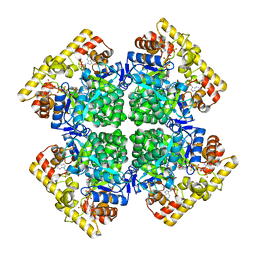 | | Crystal structure of AKRtyl-NADP(H) complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR3
 
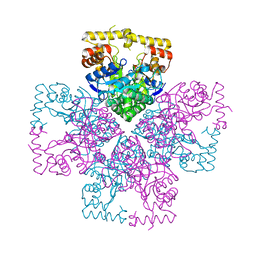 | | Crystal structure of AKRtyl-apo2 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8XR2
 
 | | Crystal structure of AKRtyl-apo1 | | Descriptor: | Aldo/keto reductase | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2024-01-06 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8KG5
 
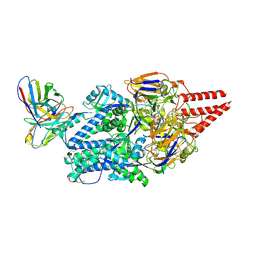 | | Prefusion RSV F Bound to Lonafarnib and D25 Fab | | Descriptor: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, D25 heavy chain, D25 light chain, ... | | Authors: | Yang, Q, Xue, B, Liu, F, Peng, W, Chen, X. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.
Signal Transduct Target Ther, 9, 2024
|
|
5Y2F
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, 9-mer peptide QTARKSTGG, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, J, Huang, Z, Song, K. | | Deposit date: | 2017-07-25 | | Release date: | 2018-11-07 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Identification of a cellularly active SIRT6 allosteric activator.
Nat. Chem. Biol., 14, 2018
|
|
8JWN
 
 | | Crystal structure of AKRtyl-NADPH complex | | Descriptor: | Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWO
 
 | |
8JWL
 
 | |
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWM
 
 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
6L6Z
 
 | |
6LFG
 
 | |
8OK4
 
 | | Variant Surface Glycoprotein VSG11wt-Oil | | Descriptor: | Variant surface glycoprotein, alpha-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Stebbins, C.E, Aresta-Branco, F, Foti, K. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8ONH
 
 | | Variant Surface Glycoprotein VSG11wt-Oil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Foti, K, Gkeka, A, Vlachou, E.P. | | Deposit date: | 2023-04-03 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8OK8
 
 | | Variant Surface Glycoprotein VSG615 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 615, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Chandra, M. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8OK5
 
 | | Variant Surface Glycoprotein VSG11 monomer with iodine | | Descriptor: | IODIDE ION, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Stebbins, C.E, Aresta-Branco, F. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8OK6
 
 | | Variant Surface Glycoprotein VSG11 two monomers | | Descriptor: | SULFATE ION, Variant surface glycoprotein, alpha-D-glucopyranose, ... | | Authors: | Gkeka, A, Vlachou, E.P, Zeelen, J.P, Stebbins, C.E. | | Deposit date: | 2023-03-27 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural classification of the variant surface glycoproteins of the African trypanosomey.
Plos Negl Trop Dis, 17, 2023
|
|
8Q0E
 
 | |
