8SAI
 
 | | Cryo-EM structure of GPR34-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2023-04-01 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5ZZ2
 
 | | Crystal structure of PDE5 in complex with inhibitor LW1634 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-05-29 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
6ACB
 
 | | Crystal structure of PDE5 in complex with inhibitor LW1805 | | Descriptor: | 3-[(2H-1,3-benzodioxol-5-yl)methyl]-8-fluoro-6-{[2-(4-methylpiperazin-1-yl)ethyl]amino}-1-(1,3-thiazol-2-yl)[1]benzopyrano[2,3-c]pyrrol-9(2H)-one, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Wu, D, Huang, Y.D, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2018-07-26 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Chromeno[2,3- c]pyrrol-9(2 H)-ones as Highly Potent, Selective, and Orally Bioavailable PDE5 Inhibitors: Structure-Activity Relationship, X-ray Crystal Structure, and Pharmacodynamic Effect on Pulmonary Arterial Hypertension.
J. Med. Chem., 61, 2018
|
|
9MQU
 
 | | CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Calcium Condition | | Descriptor: | CALCIUM ION, N~4~-(2-aminoethyl)-N~4~-methylpyrimidine-2,4-diamine, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MQS
 
 | | CryoEM Structure of the Candida albicans Group I Intron-GMP Complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-MONOPHOSPHATE, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9MQT
 
 | | CryoEM Structure of the Candida albicans Group I Intron-Compound 11 Complex under Magnesium Condition | | Descriptor: | MAGNESIUM ION, N~4~-(2-aminoethyl)-N~4~-methylpyrimidine-2,4-diamine, POTASSIUM ION, ... | | Authors: | Chung, K, Xu, L, Liu, T, Pyle, A. | | Deposit date: | 2025-01-06 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Molecular insights into de novo small-molecule recognition by an intron RNA structure.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8HZQ
 
 | | Bacillus subtilis SepF protein assembly (wild type) | | Descriptor: | Cell division protein SepF | | Authors: | Liu, W. | | Deposit date: | 2023-01-09 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular basis for curvature formation in SepF polymerization.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8HZT
 
 | |
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
7EPT
 
 | | Structural basis for the tethered peptide activation of adhesion GPCRs | | Descriptor: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ping, Y.-Q, Xiao, P, Yang, F, Zhao, R.-J, Guo, S.-C, Yan, X, Wu, X, Liebscher, I, Xu, H.E, Sun, J.-P. | | Deposit date: | 2021-04-27 | | Release date: | 2022-05-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
7EQ1
 
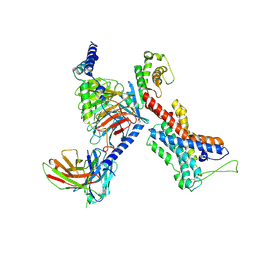 | | GPR114-Gs-scFv16 complex | | Descriptor: | Adhesion G-protein coupled receptor G5, Gs protein alpha subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ping, Y. | | Deposit date: | 2021-04-28 | | Release date: | 2022-05-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the tethered peptide activation of adhesion GPCRs.
Nature, 604, 2022
|
|
2QTF
 
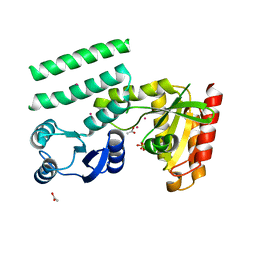 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
2QTH
 
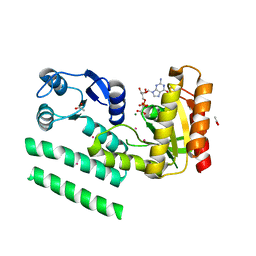 | | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus in complex with GDP | | Descriptor: | ACETATE ION, CADMIUM ION, GTP-binding protein, ... | | Authors: | Wu, H, Sun, L, Brouns, S.J, Fu, S, Rao, Z, Van der Oost, J. | | Deposit date: | 2007-08-02 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a GTP-binding protein from the hyperthermophilic archaeon Sulfolobus solfataricus.
To be Published
|
|
5DDW
 
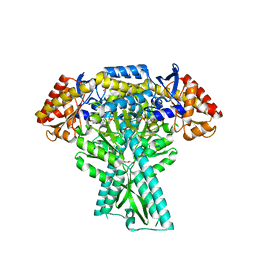 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
4RZV
 
 | | Crystal structure of the BRAF (R509H) kinase domain monomer bound to Vemurafenib | | Descriptor: | N-(3-{[5-(4-chlorophenyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]carbonyl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
8UR9
 
 | | Crystal Structure of the SARS-CoV-2 Main Protease in Complex with Compound 61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proof-of-concept studies with a computationally designed M pro inhibitor as a synergistic combination regimen alternative to Paxlovid.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3N94
 
 | | Crystal structure of human pituitary adenylate cyclase 1 Receptor-short N-terminal extracellular domain | | Descriptor: | Fusion protein of Maltose-binding periplasmic protein and pituitary adenylate cyclase 1 Receptor-short, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kumar, S, Pioszak, A.A, Swaminathan, K, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the PAC1R Extracellular Domain Unifies a Consensus Fold for Hormone Recognition by Class B G-Protein Coupled Receptors.
Plos One, 6, 2011
|
|
4RZW
 
 | | Crystal structure of BRAF (R509H) kinase domain bound to AZ628 | | Descriptor: | 3-(2-cyanopropan-2-yl)-N-{4-methyl-3-[(3-methyl-4-oxo-3,4-dihydroquinazolin-6-yl)amino]phenyl}benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Wu, Y, Gavathiotis, E. | | Deposit date: | 2014-12-24 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.493 Å) | | Cite: | An integrated model of RAF inhibitor action predicts
inhibitor activity against oncogenic BRAF signaling
Cancer Cell, 30, 2016
|
|
1RKB
 
 | | The structure of adrenal gland protein AD-004 | | Descriptor: | LITHIUM ION, Protein AD-004, SULFATE ION | | Authors: | Ren, H, Liang, Y, Bennett, M, Su, X.D. | | Deposit date: | 2003-11-21 | | Release date: | 2005-01-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human adenylate kinase 6: An adenylate kinase localized to the cell nucleus
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6P7G
 
 | | The co-crystal structure of BRAF(V600E) with PHI1 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-[(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-[4-({[2-(morpholin-4-yl)ethyl]amino}methyl)-3-(trifluoromethyl)phenyl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-06-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
4ZBN
 
 | |
6P3D
 
 | | The co-crystal structure of BRAF(V600E) with ponatinib | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, AMMONIUM ION, ... | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-05-23 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
6BL9
 
 | |
6CFR
 
 | |
6CFU
 
 | |
