8F2T
 
 | |
8F2V
 
 | |
2BCE
 
 | | CHOLESTEROL ESTERASE FROM BOS TAURUS | | Descriptor: | CHOLESTEROL ESTERASE | | Authors: | Chen, J.C.-H, Miercke, L.J.W, Krucinski, J, Starr, J.R, Saenz, G, Wang, X, Spilburg, C.A, Lange, L.G, Ellsworth, J.L, Stroud, R.M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic cholesterol esterase at 1.6 A: novel structural features involved in lipase activation.
Biochemistry, 37, 1998
|
|
1EXQ
 
 | | CRYSTAL STRUCTURE OF THE HIV-1 INTEGRASE CATALYTIC CORE DOMAIN | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL POLYPROTEIN, ... | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-05-03 | | Release date: | 2000-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EX4
 
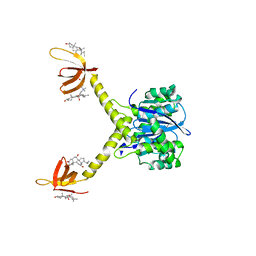 | | HIV-1 INTEGRASE CATALYTIC CORE AND C-TERMINAL DOMAIN | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, INTEGRASE | | Authors: | Chen, J.C.-H, Krucinski, J, Miercke, L.J.W, Finer-Moore, J.S, Tang, A.H, Leavitt, A.D, Stroud, R.M. | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2GVW
 
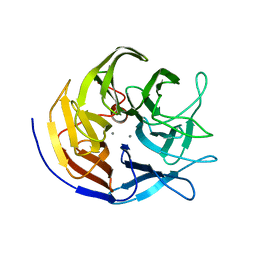 | |
2GVV
 
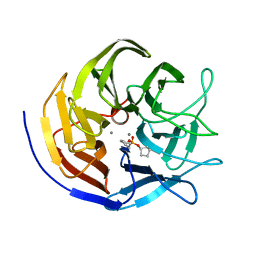 | |
2GVU
 
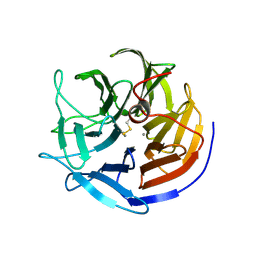 | |
3U7T
 
 | |
3LI5
 
 | | Diisopropyl fluorophosphatase (DFPase), E21Q,N120D,N175D,D229N mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3LI4
 
 | | Diisopropyl fluorophosphatase (DFPase), N120D,N175D,D229N mutant | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3LI3
 
 | | Diisopropyl fluorophosphatase (DFPase), D121E mutant | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Chen, J.C.-H. | | Deposit date: | 2010-01-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural characterization of the catalytic calcium-binding site in diisopropyl fluorophosphatase (DFPase)-Comparison with related beta-propeller enzymes.
Chem.Biol.Interact, 187, 2010
|
|
3HLI
 
 | |
3HLH
 
 | |
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | Descriptor: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | Deposit date: | 2019-12-28 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
5VG1
 
 | |
5VG0
 
 | |
3G80
 
 | |
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
8V7U
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
1KEZ
 
 | | Crystal Structure of the Macrocycle-forming Thioesterase Domain of Erythromycin Polyketide Synthase (DEBS TE) | | Descriptor: | ERYTHRONOLIDE SYNTHASE | | Authors: | Tsai, S.-C, Miercke, L.J.W, Krucinski, J, Gokhale, R, Chen, J.C.-H, Foster, P.G, Cane, D.E, Khosla, C, Stroud, R.M. | | Deposit date: | 2001-11-19 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7P6L
 
 | | Heme domain of CYP505A30, a fungal hydroxylase from Myceliophthora thermophila, bound to dodecanoic acid | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, LAURIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Opperman, D.J, Aschenbrenner, J.C, Tolmie, C, Ebrecht, A.C. | | Deposit date: | 2021-07-16 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of the fungal hydroxylase, CYP505A30, and rational transfer of mutation data from CYP102A1 to alter regioselectivity
Catalysis Science And Technology, 11, 2021
|
|
1OSF
 
 | | Human Hsp90 in complex with 17-desmethoxy-17-N,N-Dimethylaminoethylamino-Geldanamycin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 17-DESMETHOXY-17-N,N-DIMETHYLAMINOETHYLAMINO-GELDANAMYCIN, ACETIC ACID, ... | | Authors: | Jez, J.M, Chen, J.C.-H, Rastelli, G, Stroud, R.M, Santi, D.V. | | Deposit date: | 2003-03-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure and Molecular Modeling of 17-DMAG in Complex with Human Hsp90
Chem.Biol., 10, 2003
|
|
8PN6
 
 | | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus | | Descriptor: | Genome polyprotein, Serine protease subunit NS2B | | Authors: | Ni, X, Fairhead, M, Balcomb, B.H, Aschenbrenner, J.C, Ferreira, L.M, Godoy, A.S, Lithgo, R.M, MacLean, E.M, Marples, P.G, Thompson, W, Tomlinson, C.W.E, Szommer, T, Wild, C, Wright, N.D, Koekemoer, L, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-06-29 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of co-expressed NS2B-NS3 Protease from Zika Virus
To Be Published
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
