5MWE
 
 | | Complex between the Leucine Zipper (LZ, residues 490-567) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, 3,3',3''-phosphanetriyltripropanoic acid, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW9
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MVW
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Centrosomin, ... | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
5MW0
 
 | | Complex between the Leucine Zipper (LZ) and Centrosomin-motif 2 (CM2) domains of Drosophila melanogaster Centrosomin (Cnn) - L535E mutant form | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Johnson, S, Raff, J.W, Lea, S.M. | | Deposit date: | 2017-01-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
7RXB
 
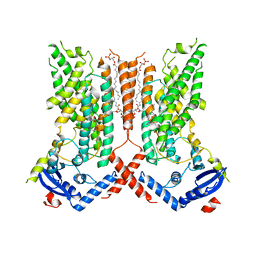 | | afTMEM16 lipid scramblase in C18 lipid nanodiscs in the absence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
7RXA
 
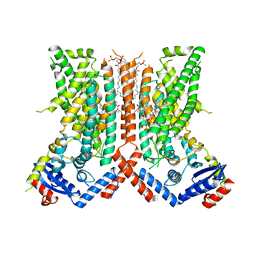 | | afTMEM16 DE/AA mutant in C14 lipid nanodiscs in the presence of Ca2+ | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, afTMEM16 lipid scramblase | | Authors: | Feng, Z, Accardi, A. | | Deposit date: | 2021-08-22 | | Release date: | 2022-05-18 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | TMEM16 scramblases thin the membrane to enable lipid scrambling.
Nat Commun, 13, 2022
|
|
1YXE
 
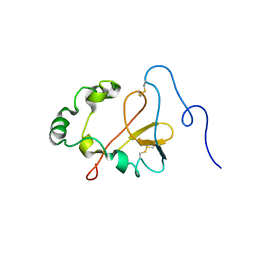 | | Structure and inter-domain interactions of domain II from the blood stage malarial protein, apical membrane antigen 1 | | Descriptor: | apical membrane antigen 1 | | Authors: | Feng, Z.P, Keizer, D.W, Stevenson, R.A, Yao, S, Babon, J.J, Murphy, V.J, Anders, R.F, Norton, R.S. | | Deposit date: | 2005-02-21 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Inter-domain Interactions of Domain II from the Blood-stage Malarial Protein, Apical Membrane Antigen 1.
J.Mol.Biol., 350, 2005
|
|
2JUS
 
 | |
2JUT
 
 | |
2JUR
 
 | |
2JUQ
 
 | |
5I7C
 
 | | Centrosomin-motif 2 (CM2) domain of Drosophila melanogaster Centrosomin (Cnn) | | Descriptor: | Centrosomin, ZINC ION | | Authors: | Feng, Z, Cottee, M.A, Johnson, S, Lea, S.M. | | Deposit date: | 2016-02-17 | | Release date: | 2017-03-08 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for Mitotic Centrosome Assembly in Flies.
Cell, 169, 2017
|
|
6PT0
 
 | | Cryo-EM structure of human cannabinoid receptor 2-Gi protein in complex with agonist WIN 55,212-2 | | Descriptor: | CHOLESTEROL, Cannabinoid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, T.H, Xing, C, Zhuang, Y, Feng, Z, Zhou, X.E, Chen, M, Wang, L, Meng, X, Xue, Y, Wang, J, Liu, H, McGuire, T, Zhao, G, Melcher, K, Zhang, C, Xu, H.E, Xie, X.Q. | | Deposit date: | 2019-07-14 | | Release date: | 2020-02-12 | | Last modified: | 2020-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM Structure of the Human Cannabinoid Receptor CB2-GiSignaling Complex.
Cell, 180, 2020
|
|
8HF8
 
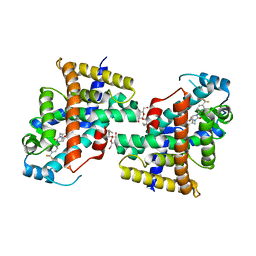 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist V1 | | Descriptor: | 2-[4-[[2,5-bis(oxidanylidene)-3-[4-(trifluoromethyl)phenyl]imidazolidin-1-yl]methyl]-2,6-dimethyl-phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta, octyl beta-D-glucopyranoside | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2022-11-09 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of the First Subnanomolar PPAR alpha / delta Dual Agonist for the Treatment of Cholestatic Liver Diseases.
J.Med.Chem., 66, 2023
|
|
7W0G
 
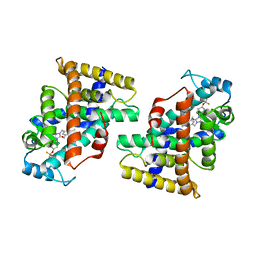 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | Descriptor: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | Deposit date: | 2021-11-18 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (2.443 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
6X5I
 
 | |
2K72
 
 | |
5DDU
 
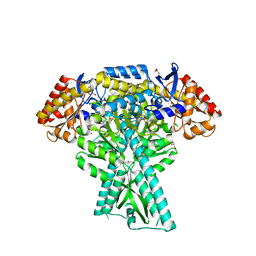 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PMP | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDS
 
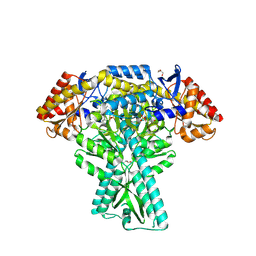 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with PLP | | Descriptor: | ACETIC ACID, CrmG, GLYCEROL, ... | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
5DDW
 
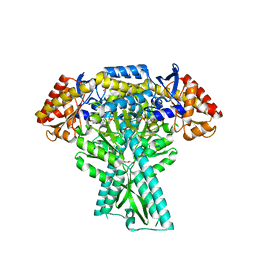 | | Crystal structure of aminotransferase CrmG from Actinoalloteichus sp. WH1-2216-6 in complex with the PMP external aldimine adduct with Caerulomycin M | | Descriptor: | CrmG, GLYCEROL, [5-hydroxy-4-({(E)-[(4-hydroxy-2,2'-bipyridin-6-yl)methylidene]amino}methyl)-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Xu, J, Feng, Z, Liu, J. | | Deposit date: | 2015-08-25 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and Structural Insights into the Aminotransferase CrmG in Caerulomycin Biosynthesis
Acs Chem.Biol., 11, 2016
|
|
7E2P
 
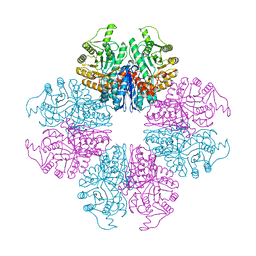 | | The Crystal Structure of Mycoplasma bovis enolase | | Descriptor: | Enolase | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Shao, G, Xiong, Q, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7XIO
 
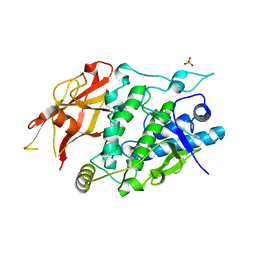 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
7E2Q
 
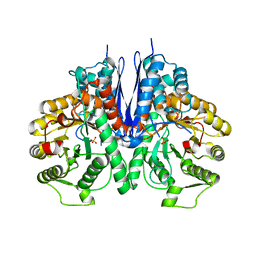 | | Crystal structure of Mycoplasma pneumoniae Enolase | | Descriptor: | Enolase, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Wang, W, Ran, T, Xiong, Q, Shao, G, Feng, Z. | | Deposit date: | 2021-02-07 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Front Mol Biosci, 8, 2022
|
|
6J36
 
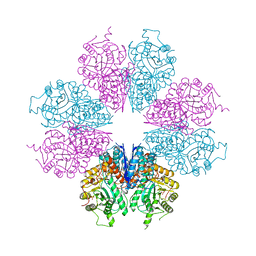 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
