7XZ6
 
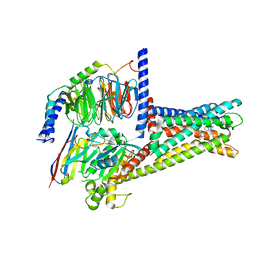 | | GPR119-Gs-APD668 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Xu, P, Huang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-06-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural identification of lysophosphatidylcholines as activating ligands for orphan receptor GPR119.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4YZW
 
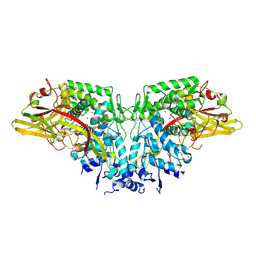 | | Crystal structure of AgPPO8 | | Descriptor: | AGAP004976-PA, COPPER (II) ION | | Authors: | Hu, Y. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a prophenoloxidase (PPO) from Anopheles gambiae provides new insights into the mechanism of PPO activation.
Bmc Biol., 14, 2016
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
7DPV
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-dihydromyricetin | | Descriptor: | (2S,3S)-3,5-dihydroxy-7-methoxy-2-(3,4,5-trihydroxyphenyl)chroman-4-one, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPP
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
7DPU
 
 | | SARS-CoV-2 3CL protease (3CLpro) in complex with 7-O-methyl-myricetin | | Descriptor: | 3C-like proteinase, 7-methoxy-3,5-bis(oxidanyl)-2-[3,4,5-tris(oxidanyl)phenyl]chromen-4-one, GLYCEROL | | Authors: | Su, H.X, Zhao, W.F, Xie, H, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease.
Nat Commun, 12, 2021
|
|
4MBS
 
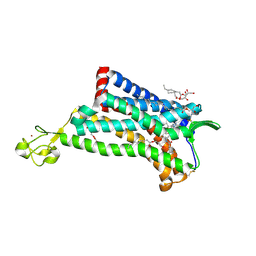 | | Crystal Structure of the CCR5 Chemokine Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]oct-8-yl}-1-phenylpropyl]cyclohexanecarboxamide, Chimera protein of C-C chemokine receptor type 5 and Rubredoxin, ... | | Authors: | Tan, Q, Zhu, Y, Han, G.W, Li, J, Fenalti, G, Liu, H, Cherezov, V, Stevens, R.C, GPCR Network (GPCR), Zhao, Q, Wu, B. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of the CCR5 chemokine receptor-HIV entry inhibitor maraviroc complex.
Science, 341, 2013
|
|
7DFG
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to favipiravir | | Descriptor: | 6-fluoro-3-oxo-4-(5-O-phosphono-beta-D-ribofuranosyl)-3,4-dihydropyrazine-2-carboxamide, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Li, Z, Zhou, Z, Yu, X. | | Deposit date: | 2020-11-08 | | Release date: | 2021-11-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
To Be Published
|
|
7DFH
 
 | |
7DOI
 
 | |
7DOK
 
 | |
8VPQ
 
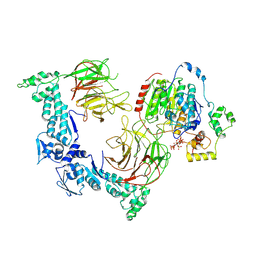 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4IPR310delinsTTYML | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Isoform 2 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-01-16 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
8VRT
 
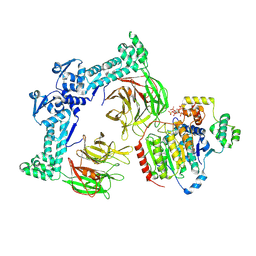 | | The structure of LSD1-CoREST-HDAC1 in complex with KBTBD4R313PRR mutant | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Xie, X, Liau, B, Zheng, N. | | Deposit date: | 2024-01-22 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Converging mechanism of UM171 and KBTBD4 neomorphic cancer mutations.
Nature, 639, 2025
|
|
7D4F
 
 | | Structure of COVID-19 RNA-dependent RNA polymerase bound to suramin | | Descriptor: | 8-(3-(3-aminobenzamido)-4-methylbenzamido)naphthalene-1,3,5-trisulfonic acid, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Li, Z, Yin, W, Zhou, Z, Yu, X, Xu, H. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for inhibition of the SARS-CoV-2 RNA polymerase by suramin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6IWI
 
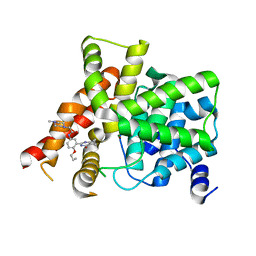 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
4P6X
 
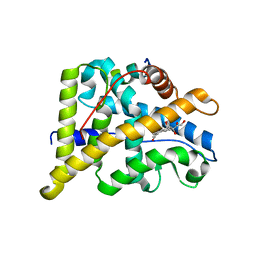 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6W
 
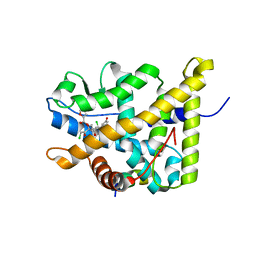 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4QKN
 
 | | Crystal structure of FTO bound to a selective inhibitor | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2014-06-07 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5.
Nucleic Acids Res., 43, 2015
|
|
8VOJ
 
 | | The Cryo-EM structure of LSD1-CoREST-HDAC1 in complex with KBTBD4 enhanced by UM171 and IP6 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Xie, X, Mao, H, Liau, B, Zheng, N. | | Deposit date: | 2024-01-15 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | UM171 glues asymmetric CRL3-HDAC1/2 assembly to degrade CoREST corepressors.
Nature, 639, 2025
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Chen, S.J, Ye, F. | | Deposit date: | 2016-08-31 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5GUV
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - R1279D | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Ye, F, Chen, S.J. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.078 Å) | | Cite: | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
8V0E
 
 | | ANK repeat of MIB1 | | Descriptor: | E3 ubiquitin-protein ligase MIB1, Unidentified MIB1 peptide | | Authors: | Cao, R, Blacklow, S.C. | | Deposit date: | 2023-11-17 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural requirements for activity of Mind bomb1 in Notch signaling.
Structure, 32, 2024
|
|
8V0D
 
 | | Ubch5B-RING3 of MIB1 fusion structure | | Descriptor: | Ubch5B-RING3 of MIB1 fusion protein, ZINC ION | | Authors: | Cao, R, Blacklow, S.C. | | Deposit date: | 2023-11-17 | | Release date: | 2024-11-27 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural requirements for activity of Mind bomb1 in Notch signaling.
Structure, 32, 2024
|
|
4HBL
 
 | | Crystal structure of AbfR of Staphylococcus epidermidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Liu, X, Sun, X, Gan, J, Lan, L, Yang, C.-G. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Oxidation-sensing Regulator AbfR Regulates Oxidative Stress Responses, Bacterial Aggregation, and Biofilm Formation in Staphylococcus epidermidis.
J.Biol.Chem., 288, 2013
|
|
7V9M
 
 | | Cryo-EM structure of the GHRH-bound human GHRHR splice variant 1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Cong, Z.T, Zhou, F.L, Zhang, C, Zou, X.Y, Zhang, H.B, Wang, Y.Z, Zhou, Q.T, Cai, X.Q, Liu, Q.F, Li, J, Shao, L.J, Mao, C.Y, Wang, X, Wu, J.H, Xia, T, Zhao, L.H, Jiang, H.L, Zhang, Y, Xu, H.E, Chen, X, Yang, D.H, Wang, M.W. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Constitutive signal bias mediated by the human GHRHR splice variant 1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
