1B3R
 
 | | RAT LIVER S-ADENOSYLHOMOCYSTEIN HYDROLASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (S-ADENOSYLHOMOCYSTEINE HYDROLASE) | | Authors: | Hu, Y, Komoto, J, Huang, Y, Takusagawa, F, Gomi, T, Ogawa, H, Takata, Y, Fujioka, M. | | Deposit date: | 1998-12-14 | | Release date: | 1998-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of S-adenosylhomocysteine hydrolase from rat liver.
Biochemistry, 38, 1999
|
|
1AMF
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE TRANSPORT PROTEIN, COMPLEXED WITH MOLYBDATE | | Descriptor: | MOLYBDATE ION, MOLYBDATE TRANSPORT PROTEIN MODA | | Authors: | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | Deposit date: | 1997-06-13 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
4W98
 
 | | Acinetobacter baumannii SDF NDK | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Hu, Y, Feng, F, Liang, H, Liu, Y. | | Deposit date: | 2014-08-27 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and Functional Characterization of Acinetobacter baumannii Nucleoside Diphosphate Kinase
Prog.Biochem.Biophys., 42, 2015
|
|
7MW6
 
 | |
7MW4
 
 | |
7MW5
 
 | |
7MW2
 
 | |
7MW3
 
 | |
8DZI
 
 | |
8DZH
 
 | |
3A9U
 
 | |
6NIL
 
 | | cryoEM structure of the truncated HIV-1 Vif/CBFbeta/A3F complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3F, Virion infectivity factor, ... | | Authors: | Hu, Y, Xiong, Y. | | Deposit date: | 2018-12-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of antagonism of human APOBEC3F by HIV-1 Vif.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1NLM
 
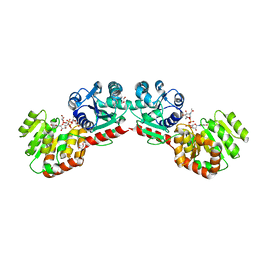 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4WBF
 
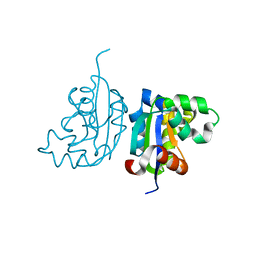 | | Acinetobacter baumannii SDF NDK | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Hu, Y, Liu, Y. | | Deposit date: | 2014-09-03 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural and Functional Characterization of Acinetobacter baumannii Nucleoside Diphosphate Kinase
Prog.Biochem.Biophys., 42, 2015
|
|
6CKH
 
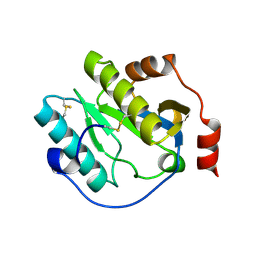 | | Manduca sexta Peptidoglycan Recognition Protein-1 | | Descriptor: | Peptidoglycan-recognition protein | | Authors: | Hu, Y. | | Deposit date: | 2018-02-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure and recognition mechanism of Manduca sexta peptidoglycan recognition protein-1.
Insect Biochem.Mol.Biol., 108, 2019
|
|
4YZW
 
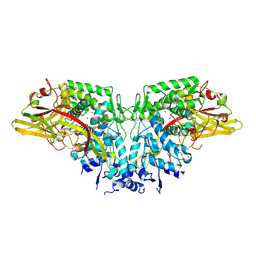 | | Crystal structure of AgPPO8 | | Descriptor: | AGAP004976-PA, COPPER (II) ION | | Authors: | Hu, Y. | | Deposit date: | 2015-03-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a prophenoloxidase (PPO) from Anopheles gambiae provides new insights into the mechanism of PPO activation.
Bmc Biol., 14, 2016
|
|
3NI2
 
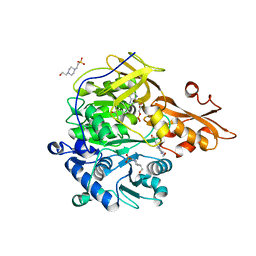 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
2HNA
 
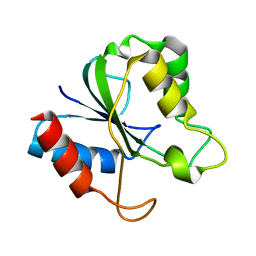 | | Solution Structure of a bacterial apo-flavodoxin | | Descriptor: | Protein mioC | | Authors: | Hu, Y, Jin, C. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
2HNB
 
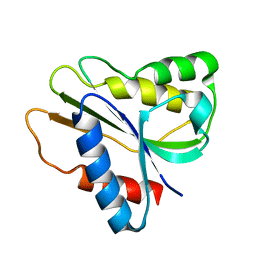 | | Solution Structure of a bacterial holo-flavodoxin | | Descriptor: | Protein mioC | | Authors: | Hu, Y, Jin, C. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structures and backbone dynamics of a flavodoxin MioC from Escherichia coli in both Apo- and Holo-forms: implications for cofactor binding and electron transfer
J.Biol.Chem., 281, 2006
|
|
7UPN
 
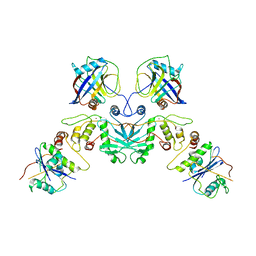 | |
1ZLG
 
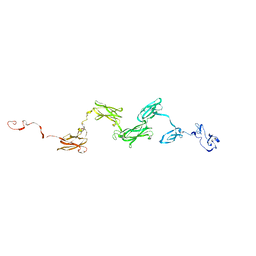 | | Solution structure of the extracellular matrix protein anosmin-1 | | Descriptor: | Anosmin 1 | | Authors: | Hu, Y, Sun, Z, Eaton, J.T, Bouloux, P.M, Perkins, S.J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-05-09 | | Last modified: | 2022-12-21 | | Method: | SOLUTION SCATTERING | | Cite: | Extended and Flexible Domain Solution Structure of the Extracellular Matrix Protein Anosmin-1 by X-ray Scattering, Analytical Ultracentrifugation and Constrained Modelling.
J.Mol.Biol., 350, 2005
|
|
1WOD
 
 | | CRYSTAL STRUCTURE OF MODA, A MOLYBDATE PROTEIN, COMPLEXED WITH TUNGSTATE | | Descriptor: | MODA, TUNGSTATE(VI)ION | | Authors: | Hu, Y, Rech, S, Gunsalus, R.P, Rees, D.C. | | Deposit date: | 1997-06-17 | | Release date: | 1997-12-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdate binding protein ModA.
Nat.Struct.Biol., 4, 1997
|
|
3A9V
 
 | |
8CMP
 
 | |
8H3F
 
 | | Cryo-EM Structure of the KBTBD2-CRL3-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
