4YCI
 
 | | non-latent pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, ... | | Authors: | Mi, L.Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1OSV
 
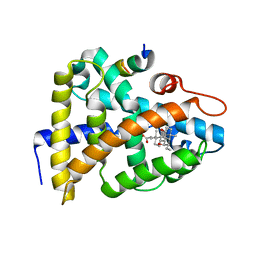 | | STRUCTURAL BASIS FOR BILE ACID BINDING AND ACTIVATION OF THE NUCLEAR RECEPTOR FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
1OT7
 
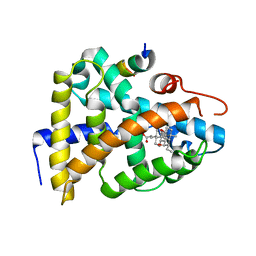 | | Structural Basis for 3-deoxy-CDCA Binding and Activation of FXR | | Descriptor: | 6-ETHYL-CHENODEOXYCHOLIC ACID, Bile Acid Receptor, ISO-URSODEOXYCHOLIC ACID, ... | | Authors: | Mi, L.Z, Devarakonda, S, Harp, J.M, Han, Q, Pellicciari, R, Willson, T.M, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2003-03-21 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Bile Acid Binding and Activation of the Nuclear Receptor FXR
Mol.Cell, 11, 2003
|
|
2AF9
 
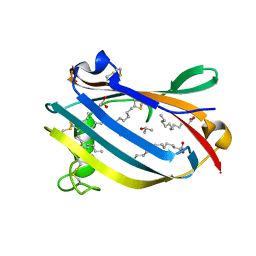 | | Crystal Structure analysis of GM2-Activator protein complexed with phosphatidylcholine | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, LAURIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG2
 
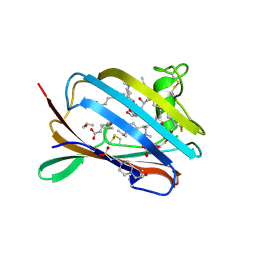 | | Crystal Structure Analysis of GM2-activator protein complexed with Phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
2AG9
 
 | | Crystal Structure of the Y137S mutant of GM2-Activator Protein | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG4
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with phosphatidylcholine | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, Ganglioside GM2 activator, ISOPROPYL ALCOHOL, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AGC
 
 | | Crystal Structure of mouse GM2- activator Protein | | Descriptor: | Ganglioside GM2 activator, LAURIC ACID, MYRISTIC ACID | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
3VDJ
 
 | | Crystal structure of circumsporozoite protein aTSR domain, R32 native form | | Descriptor: | Circumsporozoite (CS) protein | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VDL
 
 | | Crystal structure of circumsporozoite protein aTSR domain, P43212 form | | Descriptor: | Circumsporozoite (CS) protein | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VDK
 
 | | Crystal structure of circumsporozoite protein aTSR domain, R32 platinum-bound form | | Descriptor: | Circumsporozoite (CS) protein, PLATINUM (II) ION | | Authors: | Doud, M.B, Koksal, A.C, Mi, L.Z, Song, G, Lu, C, Springer, T.A. | | Deposit date: | 2012-01-05 | | Release date: | 2012-05-09 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Unexpected fold in the circumsporozoite protein target of malaria vaccines.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4YCG
 
 | | Pro-bone morphogenetic protein 9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bone Morphogenetic Protein 9 Growth Factor Domain, Bone Morphogenetic Protein 9 Prodomain, ... | | Authors: | Mi, L.-Z, Brown, C.T, Gao, Y, Tian, Y, Le, V, Walz, T, Springer, T.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of bone morphogenetic protein 9 procomplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NJP
 
 | | The Extracellular and Transmembrane Domain Interfaces in Epidermal Growth Factor Receptor Signaling | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor, Epidermal growth factor receptor, ... | | Authors: | Lu, C, Mi, L.-Z, Grey, M.J, Zhu, J, Graef, E, Yokoyama, S, Springer, T.A. | | Deposit date: | 2010-06-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Structural evidence for loose linkage between ligand binding and kinase activation in the epidermal growth factor receptor.
Mol.Cell.Biol., 30, 2010
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2B2T
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and phosphothreonine 3 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 tail | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2W
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2U
 
 | | Tandem chromodomains of human CHD1 complexed with Histone H3 Tail containing trimethyllysine 4 and dimethylarginine 2 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2Y
 
 | | Tandem chromodomains of human CHD1 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
2B2V
 
 | | Crystal structure analysis of human CHD1 chromodomains 1 and 2 bound to histone H3 resi 1-15 MeK4 | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1, Histone H3 | | Authors: | Flanagan IV, J.F, Mi, L.-Z, Chruszcz, M, Cymborowski, M, Clines, K.L, Kim, Y, Minor, W, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2005-09-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Double chromodomains cooperate to recognize the methylated histone H3 tail.
Nature, 438, 2005
|
|
4G1E
 
 | | Crystal structure of integrin alpha V beta 3 with coil-coiled tag. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Mi, L, Zhu, J, Wang, W, Luo, B, Springer, T.A. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaV Beta3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4G1M
 
 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
3V4V
 
 | | crystal structure of a4b7 headpiece complexed with Fab ACT-1 and RO0505376 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yu, Y, Zhu, J, Springer, T.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural specializations of a4b7, an Integrin that Mediates Rolling Adhesion
J.Cell Biol., 196, 2012
|
|
3V4P
 
 | | crystal structure of a4b7 headpiece complexed with Fab ACT-1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yu, Y, Zhu, J, Springer, T.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural specializations of a4b7, an Integrin that Mediates Rolling Adhesion
J.Cell Biol., 196, 2012
|
|
5I05
 
 | |
