6QWV
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Isupov, N.M, Opatowsky, Y. | | Deposit date: | 2019-03-06 | | Release date: | 2019-07-03 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural Evidence for an Octameric Ring Arrangement of SARM1.
J.Mol.Biol., 431, 2019
|
|
6KIP
 
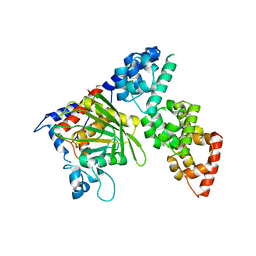 | |
6PW7
 
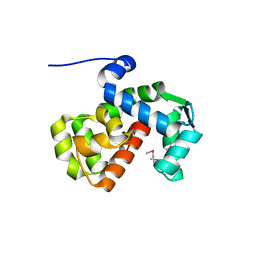 | | X-ray crystal structure of C. elegans STIM EF-SAM domain | | Descriptor: | CALCIUM ION, Stromal interaction molecule 1 | | Authors: | Enomoto, M, Nishikawa, T, Back, S.I, Ishiyama, N, Zheng, L, Stathopulos, P.B, Ikura, M. | | Deposit date: | 2019-07-22 | | Release date: | 2019-11-13 | | Last modified: | 2020-02-12 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Coordination of a Single Calcium Ion in the EF-hand Maintains the Off State of the Stromal Interaction Molecule Luminal Domain.
J.Mol.Biol., 432, 2020
|
|
6KR4
 
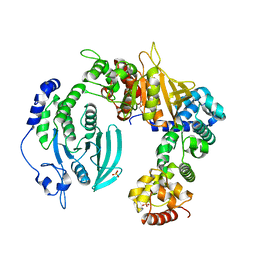 | | Crystal structure of the liprin-alpha3_SAM123/LAR_D1D2 complex | | Descriptor: | HEXAETHYLENE GLYCOL, Liprin-alpha-3, PHOSPHATE ION, ... | | Authors: | Xie, X, Liang, M, Luo, L, Wei, Z. | | Deposit date: | 2019-08-20 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of liprin-alpha-promoted LAR-RPTP clustering for modulation of phosphatase activity.
Nat Commun, 11, 2020
|
|
6WPK
 
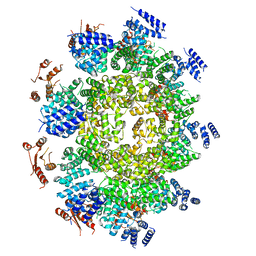 | |
6ZFX
 
 | | hSARM1 GraFix-ed | | Descriptor: | (~{E})-4-methylnon-4-enedial, NAD(+) hydrolase SARM1 | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-18 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
6ZG0
 
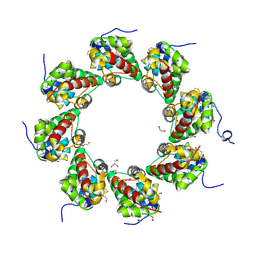 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7CM5
 
 | | Full-length Sarm1 in a self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-24 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM7
 
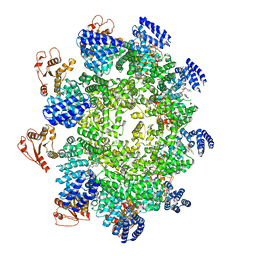 | | NAD+-bound Sarm1 E642A in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7CM6
 
 | | NAD+-bound Sarm1 in the self-inhibited state | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zhang, Z, Jiang, Y. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The NAD + -mediated self-inhibition mechanism of pro-neurodegenerative SARM1.
Nature, 588, 2020
|
|
7ANW
 
 | | hSARM1 NAD+ complex | | Descriptor: | NAD(+) hydrolase SARM1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Mim, C, Isupov, M.N, Zalk, R, Dessau, M, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-10-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
7KNQ
 
 | | SARM1 Octamer | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Shen, C, Wu, H. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-17 | | Last modified: | 2022-06-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multiple domain interfaces mediate SARM1 autoinhibition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DJT
 
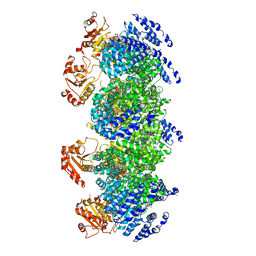 | | Human SARM1 inhibitory state bounded with inhibitor dHNN | | Descriptor: | NAD(+) hydrolase SARM1, O3-methyl O5-(2-methylpropyl) 2,6-dimethyl-4-[2-(oxidanylamino)phenyl]pyridine-3,5-dicarboxylate | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2020-11-21 | | Release date: | 2021-05-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate.
Elife, 10, 2021
|
|
7LD0
 
 | | Cryo-EM structure of ligand-free Human SARM1 | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Nanson, J.D, Gu, W, Luo, Z, Jia, X, Landsberg, M.J, Kobe, B, Ve, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARM1 is a metabolic sensor activated by an increased NMN/NAD + ratio to trigger axon degeneration.
Neuron, 109, 2021
|
|
7NAK
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7NAL
 
 | | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (ARM and SAM domains) | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1 | | Authors: | Kerry, P.S, Nanson, J.D, Adams, S, Cunnea, K, Bosanac, T, Kobe, B, Hughes, R.O, Ve, T. | | Deposit date: | 2021-06-21 | | Release date: | 2022-03-23 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules.
Mol.Cell, 82, 2022
|
|
7QG0
 
 | | Inhibitor-induced hSARM1 duplex | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Zalk, R, Kahzma, T, Guez-Haddad, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A duplex structure of SARM1 octamers stabilized by a new inhibitor.
Cell.Mol.Life Sci., 80, 2022
|
|
7Z72
 
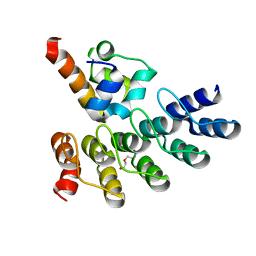 | | Crystal structure of p63 SAM in complex with darpin A5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Darpin A5, Isoform 9 of Tumor protein 63 | | Authors: | Chaikuad, A, Strubel, A, Doetsch, V, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-03-14 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
7UJN
 
 | | Structure of Human SAMHD1 with Non-Hydrolysable dGTP Analog | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | | Authors: | Huynh, K.W, Ammirati, M, Han, S. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Phosphorylation of SAMHD1 Thr592 increases C-terminal domain dynamics, tetramer dissociation and ssDNA binding kinetics.
Nucleic Acids Res., 50, 2022
|
|
8D94
 
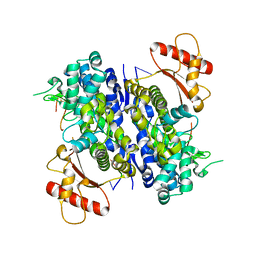 | | SAMHD1-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(P*TP*GP*T)-3'), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Hollis, T.J, Batalis, S.M. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | SAMHD1-DNA complex
To Be Published
|
|
8D9J
 
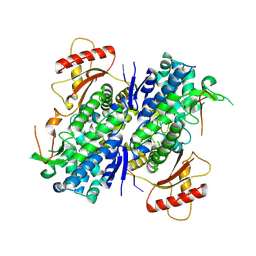 | | SAMHD1-DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*CP*AP*AP*TP*G)-3'), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Hollis, T.J, Batalis, S.M. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | SAMHD1-DNA complex
To Be Published
|
|
8ALY
 
 | | Cryo-EM structure of human tankyrase 2 SAM-PARP filament (G1032W mutant) | | Descriptor: | Poly [ADP-ribose] polymerase tankyrase-2, ZINC ION | | Authors: | Mariotti, L, Inian, O, Desfosses, A, Beuron, F, Morris, E.P, Guettler, S. | | Deposit date: | 2022-08-01 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of tankyrase activation by polymerization.
Nature, 612, 2022
|
|
8GNJ
 
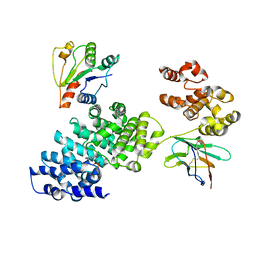 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody-C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
8GNI
 
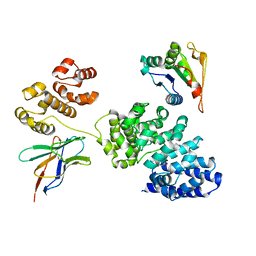 | | Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1 | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NAD(+) hydrolase SARM1, Nanobody C6 | | Authors: | Cai, Y, Zhang, H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-01-18 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | A conformation-specific nanobody targeting the nicotinamide mononucleotide-activated state of SARM1.
Nat Commun, 13, 2022
|
|
