2IWC
 
 | | Benzylpenicilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | METHICILLIN RESISTANCE MECR1 PROTEIN, OPEN FORM - PENICILLIN G | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
4L0L
 
 | | Crystal structure of P.aeruginosa PBP3 in complex with compound 4 | | Descriptor: | (6R,7S,10Z)-10-(2-amino-1,3-thiazol-4-yl)-1-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-7-formyl-13,13-dimethyl-3,9-dioxo-6-(sulfoamino)-12-oxa-2,4,8,11-tetraazatetradec-10-en-14-oic acid, Penicillin-binding protein 3 | | Authors: | Han, S, Marr, E.S. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridone-conjugated monobactam antibiotics with gram-negative activity.
J.Med.Chem., 56, 2013
|
|
4Y7P
 
 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
1NRF
 
 | | C-terminal domain of the Bacillus licheniformis BlaR penicillin-receptor | | Descriptor: | REGULATORY PROTEIN BLAR1 | | Authors: | Kerff, F, Charlier, P, Columbo, M.L, Sauvage, E, Brans, A, Frere, J.M, Joris, B, Fonze, E. | | Deposit date: | 2003-01-24 | | Release date: | 2004-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis.
Biochemistry, 42, 2003
|
|
1PYY
 
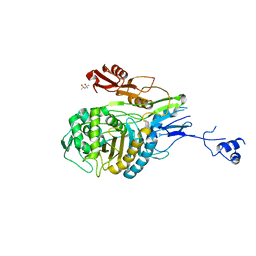 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
4P29
 
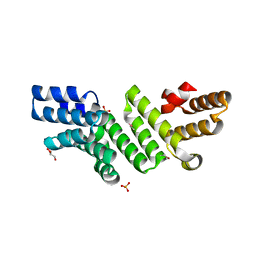 | |
4OON
 
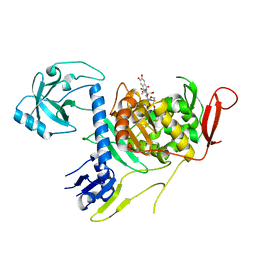 | | Crystal structure of PBP1a in complex with compound 17 ((4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid) | | Descriptor: | (4Z,8S,11E,14S)-5-(2-amino-1,3-thiazol-4-yl)-14-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-8-formyl-2-methyl-6-oxo-3,10-dioxa-4,7,11-triazatetradeca-4,11-diene-2,12,14-tricarboxylic acid, Penicillin-binding protein 1A | | Authors: | Han, S, Caspers, N, Knafels, J.D. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Siderophore receptor-mediated uptake of lactivicin analogues in gram-negative bacteria.
J.Med.Chem., 57, 2014
|
|
2QMI
 
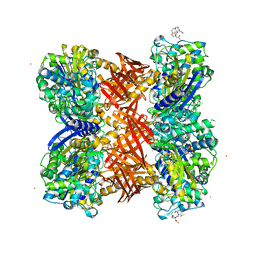 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
7JWL
 
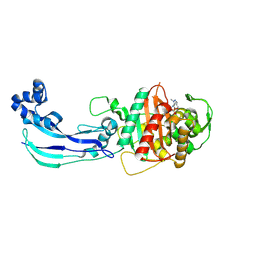 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | Descriptor: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | Deposit date: | 2020-08-25 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
2IWD
 
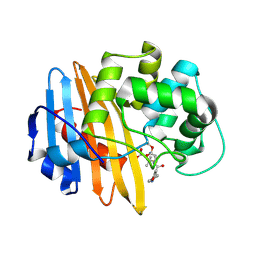 | | Oxacilloyl-acylated MecR1 extracellular antibiotic-sensor domain. | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Methicillin resistance mecR1 protein | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
2IWB
 
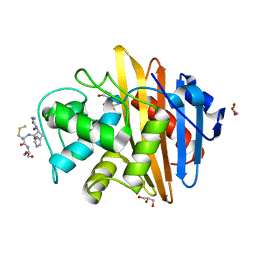 | | MecR1 unbound extracellular antibiotic-sensor domain. | | Descriptor: | GLYCEROL, METHICILLIN RESISTANCE MECR1 PROTEIN, NICKEL (II) ION, ... | | Authors: | Marrero, A, Mallorqui-Fernandez, G, Guevara, T, Garcia-Castellanos, R, Gomis-Ruth, F.X. | | Deposit date: | 2006-06-27 | | Release date: | 2006-07-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unbound and Acylated Structures of the Mecr1 Extracellular Antibiotic-Sensor Domain Provide Insights Into the Signal-Transduction System that Triggers Methicillin Resistance.
J.Mol.Biol., 361, 2006
|
|
4BL2
 
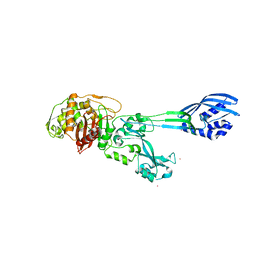 | |
1MWT
 
 | |
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4CPK
 
 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
2XK1
 
 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, SULFATE ION, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2010-07-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
2XLN
 
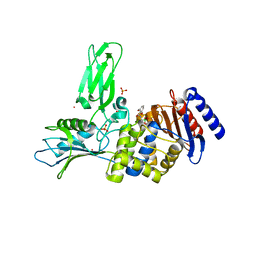 | | Crystal structure of a complex between Actinomadura R39 DD-peptidase and a boronate inhibitor | | Descriptor: | COBALT (II) ION, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE,, SULFATE ION, ... | | Authors: | Sauvage, E, Herman, R, Kerff, F, Rocaboy, M, Charlier, P. | | Deposit date: | 2010-07-21 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure Guided Development of Potent Reversibly Binding Penicillin Binding Protein Inhibitors
Acs Med.Chem.Lett., 2, 2011
|
|
6C84
 
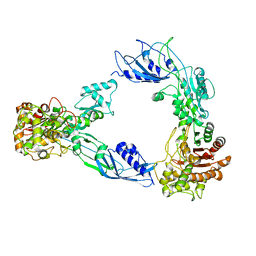 | |
7O61
 
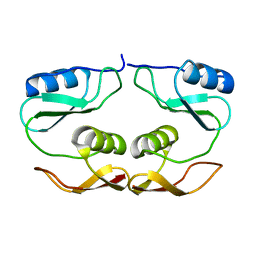 | |
3M9G
 
 | | Crystal structure of the three-PASTA-domain of a Ser/Thr kinase from Staphylococcus aureus | | Descriptor: | Protein kinase, ZINC ION | | Authors: | Paracuellos, P, Ballandras, A, Robert, X, Creze, C, Cozzone, A.J, Duclos, B, Gouet, P. | | Deposit date: | 2010-03-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extended Conformation of the 2.9-A Crystal Structure of the Three-PASTA Domain of a Ser/Thr Kinase from the Human Pathogen Staphylococcus aureus
J.Mol.Biol., 404, 2010
|
|
2OLU
 
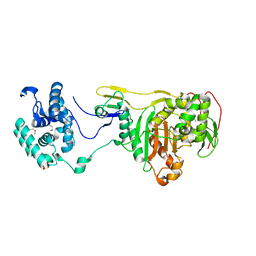 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Apoenzyme | | Descriptor: | 1,2-ETHANEDIOL, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L.H, Lim, D, Strynadka, N.C. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2OLV
 
 | | Structural Insight Into the Transglycosylation Step Of Bacterial Cell Wall Biosynthesis : Donor Ligand Complex | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 2 | | Authors: | Lovering, A.L, De Castro, L, Lim, D, Strynadka, N.C.J. | | Deposit date: | 2007-01-19 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis.
Science, 315, 2007
|
|
2D45
 
 | | Crystal structure of the MecI-mecA repressor-operator complex | | Descriptor: | 5'-D(P*TP*AP*CP*TP*AP*CP*AP*TP*AP*TP*GP*TP*AP*GP*TP*A)-3', Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Ko, T.-P, Musayev, F.N, Zhao, Q, Wang, A.H.-J, Archer, G.L. | | Deposit date: | 2005-10-09 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the MecI repressor from Staphylococcus aureus in complex with the cognate DNA operator of mec.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
6Y6Z
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 1 | | Descriptor: | GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI, ~{tert}-butyl ~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]carbamate | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion.
Chem Sci, 11, 2020
|
|
6Y6U
 
 | | Structure of Pseudomonas aeruginosa Penicillin-Binding Protein 3 (PBP3) in complex with Compound 6 | | Descriptor: | 2-(4-hydroxyphenyl)-~{N}-[(2~{S})-2-methyl-4-oxidanyl-1-oxidanylidene-pent-4-en-2-yl]ethanamide, GLYCEROL, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, D, Dowson, C.G. | | Deposit date: | 2020-02-27 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Demonstration of the utility of DOS-derived fragment libraries for rapid hit derivatisation in a multidirectional fashion
Chem Sci, 11, 2020
|
|
