6FG1
 
 | |
6FI5
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 3.2d with 14-3-3sigma | | Descriptor: | (2~{S})-2-(diphenylmethyl)pyrrolidine, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-01-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
4R67
 
 | | Human constitutive 20S proteasome in complex with carfilzomib | | Descriptor: | N-{(2S)-2-[(morpholin-4-ylacetyl)amino]-4-phenylbutanoyl}-L-leucyl-N-[(2R,3S,4S)-1,3-dihydroxy-2,6-dimethylheptan-4-yl]-L-phenylalaninamide, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-22 | | Release date: | 2015-02-11 | | Last modified: | 2015-02-18 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
6FAW
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2c-I with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{R})-[2-(2-methoxyethoxy)phenyl]-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6FCF
 
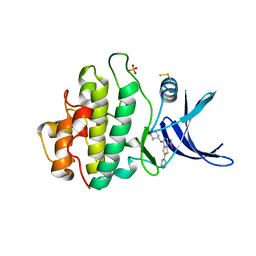 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 44 | | Descriptor: | 4-[[(2~{R},3~{S})-2-methylpiperidin-3-yl]amino]-2-phenyl-thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5YE3
 
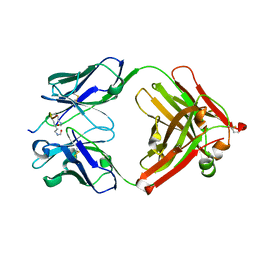 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5YKN
 
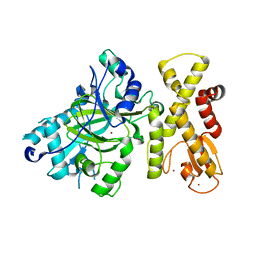 | | crystal structure of Arabidopsis thaliana JMJ14 catalytic domain | | Descriptor: | NICKEL (II) ION, Probable lysine-specific demethylase JMJ14, ZINC ION | | Authors: | Yang, Z, Du, J. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases.
Plant Cell, 30, 2018
|
|
6FBW
 
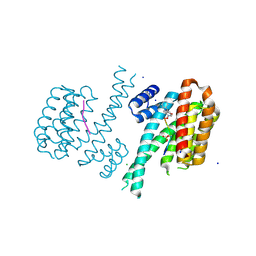 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-II with 14-3-3sigma | | Descriptor: | (2~{R})-2-[(~{S})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | Authors: | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | Deposit date: | 2017-12-20 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
6CGF
 
 | | Crystal structure of HIV-1 Y188L mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
4R3O
 
 | | Human Constitutive 20S Proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Sacchettini, J.C, Harshbarger, W.H. | | Deposit date: | 2014-08-16 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Human 20S Proteasome in Complex with Carfilzomib.
Structure, 23, 2015
|
|
6FC8
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-(3-fluorophenyl)-4-[[(3~{S})-piperidin-3-yl]amino]thieno[3,2-c]pyridine-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6FCK
 
 | | CHK1 KINASE IN COMPLEX WITH COMPOUND 13 | | Descriptor: | 2-phenyl-4-[[(3~{S})-piperidin-3-yl]amino]-1~{H}-indole-7-carboxamide, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Read, J.A, Breed, J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Adventures in Scaffold Morphing: Discovery of Fused Ring Heterocyclic Checkpoint Kinase 1 (CHK1) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6C8C
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain in complex with JHRE06 | | Descriptor: | 8-chloro-2-[(2,4-dichlorophenyl)amino]-3-(3-methylbutanoyl)-5-nitroquinolin-4(1H)-one, Chimeric protein of the Pol Kappa RIR helix and the Rev1 C-terminal domain | | Authors: | Najeeb, J, Zhou, P. | | Deposit date: | 2018-01-24 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
6FEK
 
 | |
6FF9
 
 | | Mutant R280K of human P53 | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Trovao, F.G, Gomes, A.S, Pinheiro, B, Carvalho, A.L, Romao, M.J. | | Deposit date: | 2018-01-04 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the R280K Mutant of Human p53 Explains the Loss of DNA Binding.
Int J Mol Sci, 19, 2018
|
|
6C0R
 
 | | Crystal structure of HIV-1 K103N/Y181C mutant reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C59
 
 | | Chimeric Pol kappa RIR Rev1 C-terminal domain | | Descriptor: | ACETATE ION, Chimeric Pol kappa RIR Rev1 C-terminal domain | | Authors: | Wojtaszek, J.L, Zhou, P. | | Deposit date: | 2018-01-15 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Small Molecule Targeting Mutagenic Translesion Synthesis Improves Chemotherapy.
Cell, 178, 2019
|
|
5YZ3
 
 | | Crystal structure of T2R-TTL-28 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2017-12-12 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | A Novel Microtubule Inhibitor Overcomes Multidrug Resistance in Tumors.
Cancer Res., 78, 2018
|
|
5Z2S
 
 | | Crystal structure of DUX4-HD2 domain | | Descriptor: | Double homeobox protein 4 | | Authors: | Dong, X, Zhang, W, Wu, H, Huang, J, Zhang, M, Wang, P, Zhang, H, Chen, Z, Chen, S, Meng, G. | | Deposit date: | 2018-01-03 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
6FSM
 
 | | Crystal structure of TCE-treated Thermolysin | | Descriptor: | CALCIUM ION, GLYCEROL, LYSINE, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Improved protein-crystal identification by using 2,2,2-trichloroethanol as a fluorescence enhancer.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3K8S
 
 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
6C0N
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6CGC
 
 | | Crystal structure of human 17beta-HSD type 1 in ternary complex with 2-MeO-CC-156 and NADP+ | | Descriptor: | 3-{[(9beta,14beta,16alpha,17alpha)-3,17-dihydroxy-2-methoxyestra-1,3,5(10)-trien-16-yl]methyl}benzamide, Estradiol 17-beta-dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, T, Lin, S.X. | | Deposit date: | 2018-02-19 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Combined Biophysical Chemistry Reveals a New Covalent Inhibitor with a Low-Reactivity Alkyl Halide.
J Phys Chem Lett, 9, 2018
|
|
5WA6
 
 | |
5WG9
 
 | | Crystal structure of the influenza virus PA endonuclease (E119D mutant) in complex with inhibitor 9b (SRI-30101) | | Descriptor: | 2-{(2S)-1-[3-(2-chlorophenyl)propanoyl]pyrrolidin-2-yl}-5-hydroxy-6-oxo-N-(2-phenoxyethyl)-1,6-dihydropyrimidine-4-carboxamide, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-07-13 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Protein-Structure Assisted Optimization of 4,5-Dihydroxypyrimidine-6-Carboxamide Inhibitors of Influenza Virus Endonuclease.
Sci Rep, 7, 2017
|
|
