3OOB
 
 | | Structural and functional insights of directly targeting Pin1 by Epigallocatechin-3-gallate | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Urusova, D.V, Shim, J.-H, Kim, D.-J, Jung, S.K, Zykova, T.A, Bode, A.M, Dong, Z. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Epigallocatechin-gallate suppresses tumorigenesis by directly targeting Pin1.
Cancer Prev Res (Phila), 4, 2011
|
|
1GTQ
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE, ZINC ION | | Authors: | Nar, H, Huber, R, Heizmann, C.W, Thoeny, B, Buergisser, D. | | Deposit date: | 1995-09-16 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of 6-pyruvoyl tetrahydropterin synthase, an enzyme involved in tetrahydrobiopterin biosynthesis.
EMBO J., 13, 1994
|
|
8IKH
 
 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1R)-1-(2-methoxyphenyl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8IKG
 
 | | Cryo-EM structure of human receptor with G proteins | | Descriptor: | 3-[(1S)-1-(furan-2-yl)-2-nitro-ethyl]-2-phenyl-1H-indole, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shen, S.Y, Shao, Z.H. | | Deposit date: | 2023-02-28 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based identification of a G protein-biased allosteric modulator of cannabinoid receptor CB1.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OJW
 
 | | Disulfide crosslinked cytochrome P450 reductase inactive | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH-cytochrome P450 reductase | | Authors: | Xia, C, Hamdane, D, Shen, A, Choi, V, Kasper, C, Zhang, H, Im, S.-C, Waskell, L, Kim, J.-J.P. | | Deposit date: | 2010-08-23 | | Release date: | 2011-02-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes of NADPH-Cytochrome P450 Oxidoreductase Are Essential for Catalysis and Cofactor Binding.
J.Biol.Chem., 286, 2011
|
|
8K4S
 
 | | CryoEM structure of Gq coupled MRGPRX4 with agonist DCA-3P | | Descriptor: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-12-oxidanyl-3-phosphonooxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Gs-mini-Gq chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2023-07-20 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-guided discovery of bile acid derivatives for treating liver diseases without causing itch.
Cell, 187, 2024
|
|
8JPP
 
 | | Cryo-EM structure of succinate receptor bound to succinate acid coupling MiniGsq | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
8JLZ
 
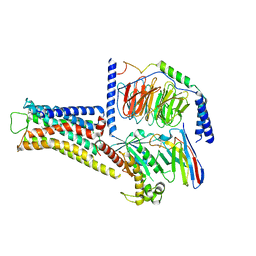 | | ST1936-5HT6R complex | | Descriptor: | 2-(5-chloranyl-2-methyl-1~{H}-indol-3-yl)-N,N-dimethyl-ethanamine, 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wen, X, Sun, J. | | Deposit date: | 2023-06-04 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insight into the selective agonist ST1936 binding of serotonin receptor 5-HT6.
Biochem.Biophys.Res.Commun., 671, 2023
|
|
6ZPL
 
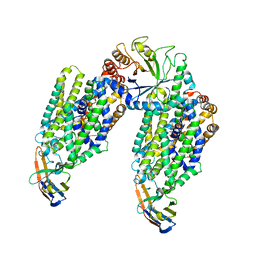 | | Inward-open structure of human glycine transporter 1 in complex with a benzoylisoindoline inhibitor, sybody Sb_GlyT1#7 and bound Na and Cl ions. | | Descriptor: | CHLORIDE ION, Endoglucanase H, SODIUM ION, ... | | Authors: | Shahsavar, A, Stohler, P, Bourenkov, G, Zimmermann, I, Siegrist, M, Guba, W, Pinard, E, Sinning, S, Seeger, M.A, Schneider, T.R, Dawson, R.J.P, Nissen, P. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.945 Å) | | Cite: | Structural insights into the inhibition of glycine reuptake.
Nature, 591, 2021
|
|
8JD6
 
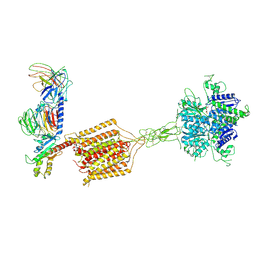 | | Cryo-EM structure of Gi1-bound metabotropic glutamate receptor mGlu4 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-3, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD5
 
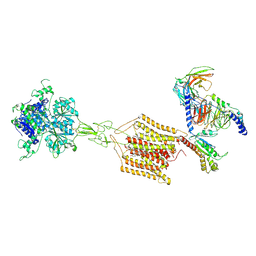 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu4 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-N-(4-methylpyrimidin-2-yl)-4-(1H-pyrazol-4-yl)-1,3-thiazol-2-amine, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JD3
 
 | | Cryo-EM structure of Gi1-bound mGlu2-mGlu3 heterodimer | | Descriptor: | 1-butyl-3-chloranyl-4-(4-phenylpiperidin-1-yl)pyridin-2-one, CHOLESTEROL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Wang, X, Wang, M, Xu, T, Feng, Y, Han, S, Lin, S, Zhao, Q, Wu, B. | | Deposit date: | 2023-05-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers.
Cell Res., 33, 2023
|
|
8JKB
 
 | | Cryo-EM structure of KCTD5 in complex with Gbeta gamma subunits | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Zheng, S, Jiang, W, Wang, W, Kong, Y. | | Deposit date: | 2023-06-01 | | Release date: | 2023-07-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural basis for the ubiquitination of G protein beta gamma subunits by KCTD5/Cullin3 E3 ligase.
Sci Adv, 9, 2023
|
|
8JR9
 
 | | Small molecule agonist (PCO371) bound to human parathyroid hormone receptor type 1 (PTH1R) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha-1, ... | | Authors: | Zhao, L, He, Q, Yuan, Q, Gu, Y, He, X, Shan, H, Li, J, Wang, K, Li, Y, Hu, W, Wu, K, Shen, J, Xu, H.E. | | Deposit date: | 2023-06-16 | | Release date: | 2023-08-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Conserved class B GPCR activation by a biased intracellular agonist.
Nature, 621, 2023
|
|
8JIL
 
 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
1DYJ
 
 | | ISOMORPHOUS CRYSTAL STRUCTURES OF ESCHERICHIA COLI DIHYDROFOLATE REDUCTASE COMPLEXED WITH FOLATE, 5-DEAZAFOLATE AND 5,10-DIDEAZATETRAHYDROFOLATE: MECHANISTIC IMPLICATIONS | | Descriptor: | 5,10-DIDEAZATETRAHYDROFOLIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reyes, V.M, Kraut, J. | | Deposit date: | 1994-10-26 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Isomorphous crystal structures of Escherichia coli dihydrofolate reductase complexed with folate, 5-deazafolate, and 5,10-dideazatetrahydrofolate: mechanistic implications.
Biochemistry, 34, 1995
|
|
8JII
 
 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIP
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JHY
 
 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
3PBG
 
 | | 6-PHOSPHO-BETA-GALACTOSIDASE FORM-C | | Descriptor: | 6-PHOSPHO-BETA-D-GALACTOSIDASE, SULFATE ION | | Authors: | Wiesmann, C, Schulz, G.E. | | Deposit date: | 1997-02-21 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures and mechanism of 6-phospho-beta-galactosidase from Lactococcus lactis.
J.Mol.Biol., 269, 1997
|
|
8JIR
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist SAR425899-bound human GLP-1R-Gs complex | | Descriptor: | Glucagon-like peptide 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JIT
 
 | | Cryo-EM structure of the GLP-1R/GCGR dual agonist MEDI0382-bound human GCGR-Gs complex | | Descriptor: | Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, L, Zhou, Q.T, Dai, A.T, Zhao, F.H, Chang, R.L, Ying, T.L, Wu, B.L, Yang, D.H, Wang, M.W, Cong, Z.T. | | Deposit date: | 2023-05-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural analysis of the dual agonism at GLP-1R and GCGR.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
