1QOW
 
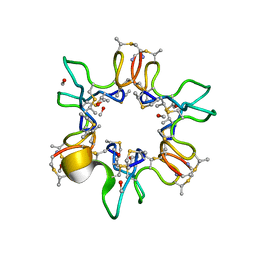 | |
2WSY
 
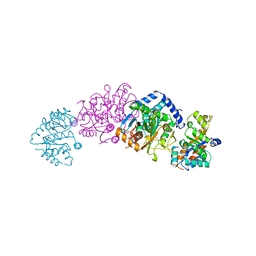 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1A50
 
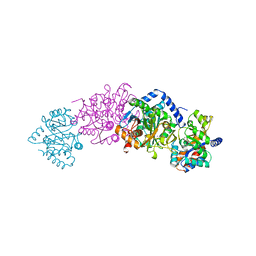 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH 5-FLUOROINDOLE PROPANOL PHOSPHATE | | Descriptor: | 5-FLUOROINDOLE PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1A5S
 
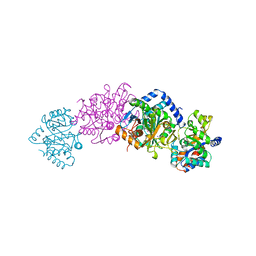 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH 5-FLUOROINDOLE PROPANOL PHOSPHATE AND L-SER BOUND AS AMINO ACRYLATE TO THE BETA SITE | | Descriptor: | 5-FLUOROINDOLE PROPANOL PHOSPHATE, PYRIDOXAL-5'-PHOSPHATE, SERINE, ... | | Authors: | Schneider, T.R, Gerhardt, E, Lee, M, Liang, P.-H, Anderson, K.S, Schlichting, I. | | Deposit date: | 1998-02-17 | | Release date: | 1999-03-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Loop closure and intersubunit communication in tryptophan synthase.
Biochemistry, 37, 1998
|
|
1HFB
 
 | |
1HAZ
 
 | | Snapshots of serine protease catalysis: (C) acyl-enzyme intermediate between porcine pancreatic elastase and human beta-casomorphin-7 jumped to pH 9 for 1 minute | | Descriptor: | BETA-CASOMORPHIN-7, CALCIUM ION, ELASTASE 1, ... | | Authors: | Wilmouth, R.C, Edman, K, Neutze, R, Wright, P.A, Clifton, I.J, Schneider, T.R, Schofield, C.J, Hajdu, J. | | Deposit date: | 2001-04-10 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Snapshots of Serine Protease Catalysis Reveal a Tetrahedral Intermediate
Nat.Struct.Biol., 8, 2001
|
|
7AF2
 
 | | Salmonella typhimurium neuraminidase mutant (D62G) | | Descriptor: | GLYCEROL, PHOSPHATE ION, Sialidase | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vimr, E.R, Garman, E.F. | | Deposit date: | 2020-09-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.792 Å) | | Cite: | Salmonella typhimurium neuraminidase mutant (D62G)
To Be Published
|
|
7AEY
 
 | | Salmonella typhimurium neuraminidase in complex with isocarba-DANA. | | Descriptor: | (3~{S},4~{S},5~{R})-4-acetamido-3-oxidanyl-5-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]cyclohexane-1-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Salinger, M.T, Kuhn, P, Laver, W.G, Pape, T, Schneider, T.R, Sheldrick, G.M, Vasella, A.T, Vimr, E.R, Vorwerk, S, Garman, E.F. | | Deposit date: | 2020-09-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Salmonella typhimurium neuraminidase in complex with isocarba-DANA.
To Be Published
|
|
353D
 
 | | CRYSTAL STRUCTURE OF DOMAIN A OF THERMUS FLAVUS 5S RRNA AND THE CONTRIBUTION OF WATER MOLECULES TO ITS STRUCTURE | | Descriptor: | RNA (5'-R(*AP*UP*CP*CP*CP*CP*CP*GP*UP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*UP*GP*CP*GP*GP*GP*GP*GP*AP*U)-3') | | Authors: | Betzel, C, Lorenz, S, Furste, J.P, Bald, R, Zhang, M, Schneider, T.R, Wilson, K.S, Erdmann, V.A. | | Deposit date: | 1997-09-29 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure.
FEBS Lett., 351, 1994
|
|
2J8T
 
 | | Human aldose reductase in complex with NADP and citrate at 0.82 angstrom | | Descriptor: | ALDO-KETO REDUCTASE FAMILY 1, MEMBER B1, CITRATE ANION, ... | | Authors: | Biadene, M, Hazemann, I, Cousido, A, Ginell, S, Sheldrick, G.M, Podjarny, A, Schneider, T.R. | | Deposit date: | 2006-10-27 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.82 Å) | | Cite: | The Atomic Resolution Structure of Human Aldose Reductase Reveals that Rearrangement of a Bound Ligand Allows the Opening of the Safety-Belt Loop.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1W7Z
 
 | | Crystal structure of the free (uncomplexed) Ecballium elaterium trypsin inhibitor (EETI-II) | | Descriptor: | FORMIC ACID, SODIUM ION, TRYPSIN INHIBITOR II | | Authors: | Kraetzner, R, Debreczeni, J.E, Pape, T, Kolmar, H, Schneider, T.R, Uson, I, Scheldrick, G.M. | | Deposit date: | 2004-09-14 | | Release date: | 2005-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of Ecballium Elaterium Trypsin Inhibitor II (Eeti-II): A Rigid Molecular Scaffold
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5FTP
 
 | | sulfur SAD phasing of Cdc23Nterm: data collection with a tailored X- ray beam size at 2.69 A wavelength (4.6 keV) | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 8 | | Authors: | Cianci, M, Groves, M.R, Barford, D, Schneider, T.R. | | Deposit date: | 2016-01-14 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Data Collection with a Tailored X-Ray Beam Size at 2.69 A Wavelength (4.6 Kev): Sulfur Sad Phasing of Cdc23Nterm
Acta Crystallogr.,Sect.D, 72, 2016
|
|
5AHS
 
 | | 3-Sulfinopropionyl-Coenzyme A (3SP-CoA) desulfinase from Advenella mimgardefordensis DPN7T: holo crystal structure with the substrate analog succinyl-CoA | | Descriptor: | ACYL-COA DEHYDROGENASE, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1OFO
 
 | | Crystal Structure of the Tyrosine Regulated 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae in Complex with 2-Phosphoglycolate | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Koenig, V, Pfeil, A, Heinrich, G, Braus, G, Schneider, T.R. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate and Metal Complexes of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism.
J.Mol.Biol., 337, 2004
|
|
1OFB
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH MANGANESE(II) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Koenig, V, Pfeil, A, Heinrich, G, Braus, G.H, Schneider, T.R. | | Deposit date: | 2003-04-09 | | Release date: | 2004-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Substrate and Metal Complexes of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism
J.Mol.Biol., 337, 2004
|
|
1OFP
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Koenig, V, Pfeil, A, Heinrich, G, Braus, G.H, Schneider, T.R. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate and Metal Complexes of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism
J.Mol.Biol., 337, 2004
|
|
1OFQ
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE-REGULATED 3-DEOXY-D-ARABINO-HEPTULOSONATE-7-PHOSPHATE SYNTHASE FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH MANGANESE(II) | | Descriptor: | MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Koenig, V, Pfeil, A, Heinrich, G, Braus, G.H, Schneider, T.R. | | Deposit date: | 2003-04-17 | | Release date: | 2004-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate and Metal Complexes of 3-Deoxy-D-Arabino-Heptulosonate-7-Phosphate Synthase from Saccharomyces Cerevisiae Provide New Insights Into the Catalytic Mechanism
J.Mol.Biol., 337, 2004
|
|
5AF7
 
 | | 3-Sulfinopropionyl-coenzyme A (3SP-CoA) desulfinase from Advenella mimigardefordensis DPN7T: crystal structure and function of a desulfinase with an acyl-CoA dehydrogenase fold. Native crystal structure | | Descriptor: | ACYL-COA DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cianci, M, Schuermann, M, Meijers, R, Schneider, T.R, Steinbuechel, A. | | Deposit date: | 2015-01-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | 3-Sulfinopropionyl-Coenzyme a (3Sp-Coa) Desulfinase from Advenella Mimigardefordensis Dpn7(T): Crystal Structure and Function of a Desulfinase with an Acyl-Coa Dehydrogenase Fold.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1L2H
 
 | | Crystal structure of Interleukin 1-beta F42W/W120F mutant | | Descriptor: | Interleukin 1-beta | | Authors: | Rudolph, M.G, Kelker, M.S, Schneider, T.R, Yeates, T.O, Oseroff, V, Heidary, D.K, Jennings, P.A, Wilson, I.A. | | Deposit date: | 2002-02-21 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Use of multiple anomalous dispersion to phase highly merohedrally twinned crystals of interleukin-1beta.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4BLG
 
 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
4C1P
 
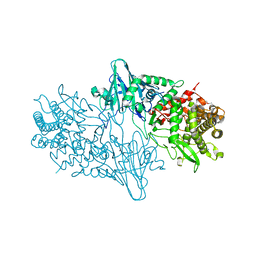 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | Descriptor: | BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | Deposit date: | 2013-08-13 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C1O
 
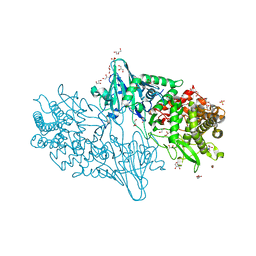 | | Geobacillus thermoglucosidasius GH family 52 xylosidase | | Descriptor: | 1,2-ETHANEDIOL, BETA-XYLOSIDASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Espina, G, Eley, K, Schneider, T.R, Crennell, S.J, Danson, M.J. | | Deposit date: | 2013-08-13 | | Release date: | 2014-05-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Novel Beta-Xylosidase Structure from Geobacillus Thermoglucosidasius: The First Crystal Structure of a Glycoside Hydrolase Family Gh52 Enzyme Reveals Unpredicted Similarity to Other Glycoside Hydrolase Folds
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2C7N
 
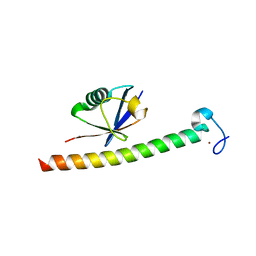 | | Human Rabex-5 residues 1-74 in complex with Ubiquitin | | Descriptor: | RAB GUANINE NUCLEOTIDE EXCHANGE FACTOR 1, UBIQUITIN, ZINC ION | | Authors: | Penengo, L, Mapelli, M, Murachelli, A.G, Confalioneri, S, Magri, L, Musacchio, A, Di Fiore, P.P, Polo, S, Schneider, T.R. | | Deposit date: | 2005-11-25 | | Release date: | 2006-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ubiquitin Binding Domains of Rabex-5 Reveals Two Modes of Interaction with Ubiquitin.
Cell(Cambridge,Mass.), 124, 2006
|
|
2BFY
 
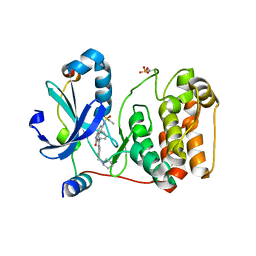 | | Complex of Aurora-B with INCENP and Hesperadin. | | Descriptor: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A, N-[2-OXO-3-((E)-PHENYL{[4-(PIPERIDIN-1-YLMETHYL)PHENYL]IMINO}METHYL)-2,6-DIHYDRO-1H-INDOL-5-YL]ETHANESULFONAMIDE | | Authors: | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | Deposit date: | 2004-12-15 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
2BFX
 
 | | Mechanism of Aurora-B activation by INCENP and inhibition by Hesperadin. | | Descriptor: | AURORA KINASE B-A, INNER CENTROMERE PROTEIN A | | Authors: | Sessa, F, Mapelli, M, Ciferri, C, Tarricone, C, Areces, L.B, Schneider, T.R, Stukenberg, P.T, Musacchio, A. | | Deposit date: | 2004-12-15 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Aurora B Activation by Incenp and Inhibition by Hesperadin
Mol.Cell, 18, 2005
|
|
