6T2E
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Stapled peptide GAR300-Gm | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2F
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[3-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, E3 ubiquitin-protein ligase Mdm2, MDM2 in complex with GAR300-Am | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6T2D
 
 | | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions | | Descriptor: | 2-(methylamino)-~{N}-[[4-[[2-(methylamino)ethanoylamino]methyl]phenyl]methyl]ethanamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Groves, R.M, Ali, M.A, Atmaj, J, van Oosterwijk, N, Domling, A, Rivera, G.D, Ricardo, G.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Multicomponent Peptide Stapling as a Diversity-Driven Tool for the Development of Inhibitors of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3G03
 
 | |
4ERF
 
 | | crystal structure of MDM2 (17-111) in complex with compound 29 (AM-8553) | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, {(3R,5R,6S)-5-(3-chlorophenyl)-6-(4-chlorophenyl)-1-[(2S,3S)-2-hydroxypentan-3-yl]-3-methyl-2-oxopiperidin-3-yl}acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
4DIJ
 
 | |
3EQS
 
 | |
4ERE
 
 | | crystal structure of MDM2 (17-111) in complex with compound 23 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(3R,5R,6S)-1-[(2S)-1-tert-butoxy-1-oxobutan-2-yl]-5-(3-chlorophenyl)-6-(4-chlorophenyl)-2-oxopiperidin-3-yl]acetic acid | | Authors: | Huang, X. | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of novel inhibitors of the MDM2-p53 interaction.
J.Med.Chem., 55, 2012
|
|
2MPS
 
 | |
2M86
 
 | | Solution structure of Hdm2 with engineered cyclotide | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, MCo-PMI | | Authors: | Majumder, S, Ji, Y, Millard, M, Borra, R, Bi, T, Elnagar, A.Y, Neamati, N, Camarero, J.A. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | In Vivo Activation of the p53 Tumor Suppressor Pathway by an Engineered Cyclotide.
J.Am.Chem.Soc., 135, 2013
|
|
3LBL
 
 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
6V4E
 
 | | Crystal Structure Analysis of Zebra Fish MDM | | Descriptor: | Protein Mdm4, Stapled peptide QSQQTF(0EH)NLWRLL(MK8)QN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
3LNZ
 
 | |
3LNJ
 
 | |
6V4G
 
 | | Crystal Structure Analysis of Zebra fish MDMX | | Descriptor: | Protein Mdm4, SULFATE ION, Stapled peptide QSQQTF(0EH)NLWRLE(MK8)QN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
3VBG
 
 | | Structure of hDM2 with Dimer Inducing Indolyl Hydantoin RO-2443 | | Descriptor: | (5Z)-5-[(6-chloro-7-methyl-1H-indol-3-yl)methylidene]-3-(3,4-difluorobenzyl)imidazolidine-2,4-dione, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Lukacs, C.M, Janson, C.A, Graves, B.J. | | Deposit date: | 2012-01-02 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Activation of the p53 pathway by small-molecule-induced MDM2 and MDMX dimerization.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XXK
 
 | |
3VZV
 
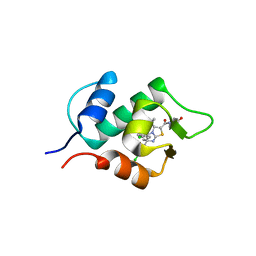 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | 1-{[(5R,6S)-5,6-bis(4-chlorophenyl)-6-methyl-3-(propan-2-yl)-5,6-dihydroimidazo[2,1-b][1,3]thiazol-2-yl]carbonyl}-N,N-dimethyl-L-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2012-10-16 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6Q9L
 
 | |
5VK0
 
 | |
6Q9O
 
 | |
6Q9H
 
 | |
7NA3
 
 | | HDM2 in complex with compound 62 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(2S)-1-methoxypropan-2-yl]-3-{(1R)-1-[(1r,4R)-4-methylcyclohexyl]ethyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
7NA2
 
 | | HDM2 in complex with compound 56 | | Descriptor: | 3-[4-(5-chloropyridin-3-yl)-2-[(4aR,7aR)-hexahydrocyclopenta[b][1,4]oxazin-4(4aH)-yl]-3-{[(1r,4R)-4-methylcyclohexyl]methyl}-3H-imidazo[4,5-c]pyridin-6-yl]-1,2,4-oxadiazol-5(4H)-one, Isoform 11 of E3 ubiquitin-protein ligase Mdm2 | | Authors: | Scapin, G. | | Deposit date: | 2021-06-19 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of MK-4688 : an Efficient Inhibitor of the HDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
