1YYY
 
 | | Trypsin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3S)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, CALCIUM ION, TRYPSIN | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-06-03 | | Release date: | 1999-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
7SPO
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with VNAR 3B4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2021-11-02 | | Release date: | 2022-01-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography.
Nat Commun, 12, 2021
|
|
7Z2W
 
 | | Escherichia coli periplasmic phytase AppA D304A,T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2T
 
 | | Escherichia coli periplasmic phytase AppA D304A mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7YXM
 
 | | Benzoylsuccinyl-CoA thiolase with coenzyme A | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Benzoylsuccinyl-CoA thiolase subunit, ... | | Authors: | Ermler, U, Heider, J, Weidenweber, S. | | Deposit date: | 2022-02-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Febs J., 289, 2022
|
|
7SB7
 
 | | Crystal structure of T. brucei hypoxanthine guanine phosphoribosyltransferase in complex with (4S,7S)-7-hydroxy-4-((guanin-9-yl)methyl)-2,5-dioxaheptan-1,7-diphosphonate | | Descriptor: | ({(2S)-3-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-2-[(2S)-2-hydroxy-2-phosphonoethoxy]propoxy}methyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Guddat, L.W, Keough, D.T. | | Deposit date: | 2021-09-24 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.64716625 Å) | | Cite: | Stereo-Defined Acyclic Nucleoside Phosphonates are Selective and Potent Inhibitors of Parasite 6-Oxopurine Phosphoribosyltransferases.
J.Med.Chem., 65, 2022
|
|
7SKQ
 
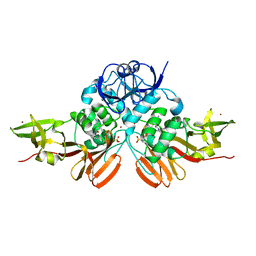 | | BtSCoV-Rf1.2004 Papain-Like protease bound to the non-covalent inhibitor GRL-0617 | | Descriptor: | 3C-like proteinase, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ZINC ION | | Authors: | Freitas, B, Durie, I, Shepard, J, O'Boyle, B, Enos, S, Pegan, S.D. | | Deposit date: | 2021-10-21 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Exploring Noncovalent Protease Inhibitors for the Treatment of Severe Acute Respiratory Syndrome and Severe Acute Respiratory Syndrome-Like Coronaviruses.
Acs Infect Dis., 8, 2022
|
|
4NLD
 
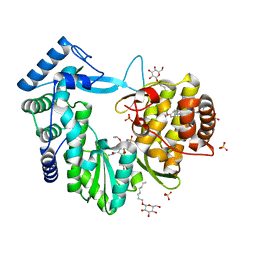 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with BMS-791325 also known as (1aR,12bS)-8-cyclohexyl-n-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide and 2-(4-fluorophenyl)-n-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide | | Descriptor: | (1aR,12bS)-8-cyclohexyl-N-(dimethylsulfamoyl)-11-methoxy-1a-{[(1R,5S)-3-methyl-3,8-diazabicyclo[3.2.1]oct-8-yl]carbonyl}-1,1a,2,12b-tetrahydrocyclopropa[d]indolo[2,1-a][2]benzazepine-5-carboxamide, 2-(4-fluorophenyl)-N-methyl-6-[(methylsulfonyl)amino]-5-(propan-2-yloxy)-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Preclinical Characterization of the Cyclopropylindolobenzazepine BMS-791325, A Potent Allosteric Inhibitor of the Hepatitis C Virus NS5B Polymerase.
J.Med.Chem., 57, 2014
|
|
7Z43
 
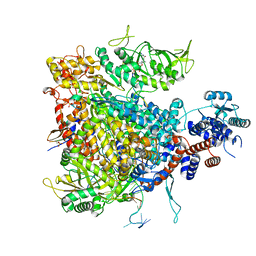 | |
7Z60
 
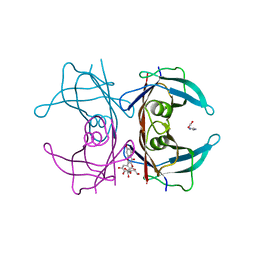 | | Human transthyretin (TTR) complexed with Quercetin 3-O-beta-D-galactoside | | Descriptor: | 1,2-ETHANEDIOL, 2-[3,4-bis(oxidanyl)phenyl]-3-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5,7-bis(oxidanyl)chromen-4-one, IMIDAZOLE, ... | | Authors: | Ciccone, L, Braca, A, Orlandini, E, Nencetti, S. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-18 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization
Crystals, 12, 2022
|
|
1ZRF
 
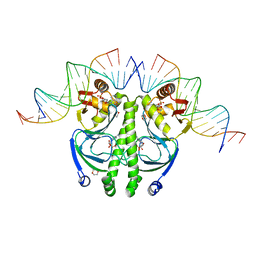 | | 4 crystal structures of CAP-DNA with all base-pair substitutions at position 6, CAP-[6C;17G]ICAP38 DNA | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 5'-D(*AP*TP*TP*TP*CP*GP*AP*AP*AP*AP*AP*TP*GP*CP*GP*AP*T)-3', 5'-D(*CP*TP*AP*GP*AP*TP*CP*GP*CP*AP*TP*TP*TP*TP*TP*CP*GP*AP*AP*AP*T)-3', ... | | Authors: | Berman, H.M, Napoli, A.A. | | Deposit date: | 2005-05-19 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: recognition of pyrimidine-purine and purine-purine steps.
J.Mol.Biol., 357, 2006
|
|
7S0S
 
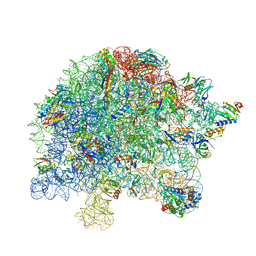 | | M. tuberculosis ribosomal RNA methyltransferase TlyA bound to M. smegmatis 50S ribosomal subunit | | Descriptor: | 16S/23S rRNA (Cytidine-2'-O)-methyltransferase TlyA, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Laughlin, Z.T, Dunham, C.M, Conn, G.L. | | Deposit date: | 2021-08-31 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | 50S subunit recognition and modification by the Mycobacterium tuberculosis ribosomal RNA methyltransferase TlyA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
7RAN
 
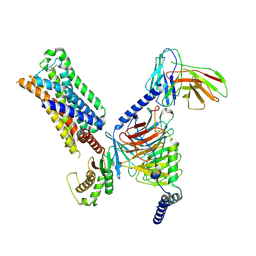 | | 5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | (3R)-3-methyl-5-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,3,6-tetrahydropyridin-1-ium, 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2021-07-02 | | Release date: | 2022-07-06 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Nature, 610, 2022
|
|
6D6U
 
 | | Human GABA-A receptor alpha1-beta2-gamma2 subtype in complex with GABA and flumazenil, conformation A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Noviello, C.M, Teng, J, Walsh Jr, R.M, Kim, J.J, Hibbs, R.E. | | Deposit date: | 2018-04-22 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of a human synaptic GABAAreceptor.
Nature, 559, 2018
|
|
7RKK
 
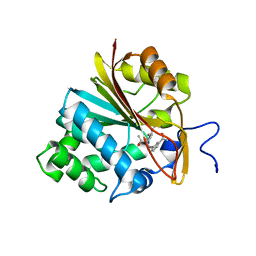 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with II399 (C2 space group) | | Descriptor: | 3-[3-(acetyl{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)prop-1-yn-1-yl]benzamide, NNMT protein | | Authors: | Yadav, R, Noinaj, N, Iyamu, I.D, Huang, R. | | Deposit date: | 2021-07-22 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Exploring Unconventional SAM Analogues To Build Cell-Potent Bisubstrate Inhibitors for Nicotinamide N-Methyltransferase.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y1Q
 
 | | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | | Descriptor: | Isoform 2 of Basigin, Monocarboxylate transporter 1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.03 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2
To Be Published
|
|
7RN2
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with SJ001010551-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[(6S,10S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-[(pyridin-2-yl)methyl]acetamide, Bromodomain-containing protein 4 | | Authors: | Stachowski, T.R, Fischer, M. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | From PROTAC to inhibitor: Structure-guided discovery of potent and orally bioavailable BET inhibitors.
Eur.J.Med.Chem., 251, 2023
|
|
7Y3H
 
 | | Crystal Structure of Diels-Alderase ApiI in complex with SAM and product | | Descriptor: | 1,2-ETHANEDIOL, 3-[[(1R,2R,4aS,8aR)-2-methyl-1,2,4a,5,6,7,8,8a-octahydronaphthalen-1-yl]carbonyl]-5-(4-hydroxyphenyl)-4-oxidanyl-1H-pyridin-2-one, ApiI, ... | | Authors: | Zhou, J.H, Lu, J.Y. | | Deposit date: | 2022-06-10 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of Diels-Alderase ApiI in complex with SAM and product
To Be Published
|
|
7Y4G
 
 | | sit-bound btDPP4 | | Descriptor: | (2R)-4-OXO-4-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-1-(2,4,5-TRIFLUOROPHENYL)BUTAN-2-A MINE, btDPP4 | | Authors: | Hang, J, Jiang, C, Wang, K, Zhang, Z, Guo, F, Liu, J, Wang, G, Lei, X, Gonzalez, F, Qiao, J. | | Deposit date: | 2022-06-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target.
Science, 381, 2023
|
|
4N4W
 
 | | Structure of the human smoothened receptor in complex with SANT-1. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (E)-N-(4-benzylpiperazin-1-yl)-1-(3,5-dimethyl-1-phenyl-1H-pyrazol-4-yl)methanimine, Cytochrome b(562),Smoothened homolog, ... | | Authors: | Wang, C, Wu, H, Han, G.W, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs.
Nat Commun, 5, 2014
|
|
8PMR
 
 | | NADase from Aspergillus fumigatus with mutated calcium binding motif (D219A/E220A) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
7YIY
 
 | | Cryo-EM structure of SPT-ORMDL3 complex | | Descriptor: | N-[(2S,3R,4E)-1,3-dihydroxyoctadec-4-en-2-yl]tetracosanamide, ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Xie, T, Liu, P, Gong, X. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ceramide sensing by human SPT-ORMDL complex for establishing sphingolipid homeostasis.
Nat Commun, 14, 2023
|
|
7RD2
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 2 | | Descriptor: | (2R,3R,4R,5S)-1-{[4-({4-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-2-nitroanilino}methyl)phenyl]methyl}-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2021-07-09 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
