2QPT
 
 | |
5NJ8
 
 | |
1SRQ
 
 | | Crystal Structure of the Rap1GAP catalytic domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GTPase-activating protein 1, SULFATE ION | | Authors: | Daumke, O, Weyand, M, Chakrabarti, P.P, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 2004-03-23 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The GTPase activating protein Rap1GAP uses a catalytic asparagine
Nature, 429, 2004
|
|
7SOX
 
 | |
7PV1
 
 | |
7PV0
 
 | | Crystal structure of a Mic60-Mic19 fusion protein | | Descriptor: | MICOS complex subunit MIC60,MICOS complex subunit MIC60-MIC19,Mic60-Mic19, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Funck, K, Bock-Bierbaum, T, Daumke, O. | | Deposit date: | 2021-10-01 | | Release date: | 2022-09-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into crista junction formation by the Mic60-Mic19 complex.
Sci Adv, 8, 2022
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6GMA
 
 | |
3LJB
 
 | |
7PUZ
 
 | |
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
8Q1X
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
8R1A
 
 | | Model of the membrane-bound GBP1 oligomer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate binding protein 1, ... | | Authors: | Weismehl, M, Chu, X, Kutsch, M, Lauterjung, P, Herrmann, C, Kudryashev, M, Daumke, O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (26.799999 Å) | | Cite: | Structural insights into the activation mechanism of antimicrobial GBP1.
Embo J., 43, 2024
|
|
5FPH
 
 | | The GTPase domains of the immunity-related Irga6 dimerize in a parallel head-to-head fashion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INTERFERON-INDUCIBLE GTPASE 1, MAGNESIUM ION, ... | | Authors: | Schulte, K, Pawlowski, N, Faelber, K, Froehlich, C, Howard, J, Daumke, O. | | Deposit date: | 2015-11-30 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Immunity-Related Gtpase Irga6 Dimerizes in a Parallel Head-to-Head Fashion.
Bmc Biol., 14, 2016
|
|
8Q1S
 
 | | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein epsilon, BROMIDE ION, ... | | Authors: | Roske, Y, Daumke, O, Rrustemi, T, Selbach, M. | | Deposit date: | 2023-08-01 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Pathogenic mutations of human phosphorylation sites affect protein-protein interactions.
Nat Commun, 15, 2024
|
|
7Z74
 
 | | PI3KC2a core in complex with PITCOIN2 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ~{N}-[4-(3-hydroxyphenyl)-1,3-thiazol-2-yl]-2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-ethanamide | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
7Z75
 
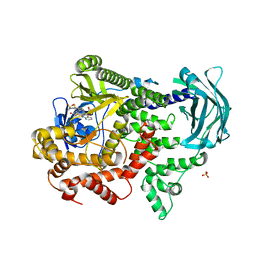 | | PI3KC2a core in complex with PITCOIN3 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
8A9I
 
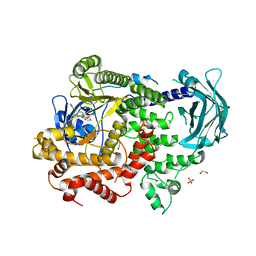 | | PI3KC2a core in complex with PITCOIN1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-~{N}-(1,3-thiazol-2-yl)ethanamide, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-06-28 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
3SZR
 
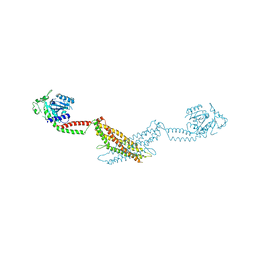 | | Crystal structure of modified nucleotide-free human MxA | | Descriptor: | Interferon-induced GTP-binding protein Mx1 | | Authors: | Gao, S, Daumke, O. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of myxovirus resistance protein a reveals intra- and intermolecular domain interactions required for the antiviral function.
Immunity, 35, 2011
|
|
7BI2
 
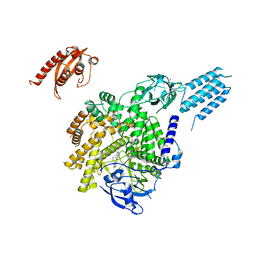 | | PI3KC2aDeltaN and DeltaC-C2 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-aminopyridin-3-yl)-1-[3-(trifluoromethyl)phenyl]benzo[h][1,6]naphthyridin-2(1H)-one, IODIDE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2021-01-12 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of phosphatidylinositol 3-kinase C2 alpha function.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5MTV
 
 | | Active structure of EHD4 complexed with ATP-gamma-S | | Descriptor: | EH domain-containing protein 4, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-10 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MVF
 
 | | Active structure of EHD4 complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EH domain-containing protein 4, MAGNESIUM ION | | Authors: | Melo, A.A, Daumke, O. | | Deposit date: | 2017-01-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.268 Å) | | Cite: | Structural insights into the activation mechanism of dynamin-like EHD ATPases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1YE8
 
 | | Crystal Structure of THEP1 from the hyperthermophile Aquifex aeolicus | | Descriptor: | Hypothetical UPF0334 kinase-like protein AQ_1292, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rossbach, M, Daumke, O, Klinger, C, Wittinghofer, A, Kaufmann, M. | | Deposit date: | 2004-12-28 | | Release date: | 2005-03-29 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of THEP1 from the hyperthermophile Aquifex aeolicus: a variation of the RecA fold
BMC Struct.Biol., 5, 2005
|
|
6YGC
 
 | | Crystal structure of the NatC complex bound to Arl3 peptide and CoA | | Descriptor: | ADP-ribosylation factor-like protein 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6YGB
 
 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
