6HGI
 
 | |
6HGL
 
 | |
6HGK
 
 | |
6HGD
 
 | |
6HGG
 
 | | Crystal structure of Alpha1-antichymotrypsin variant NewBG-III: a new binding globulin in complex with cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, 1,2-ETHANEDIOL, Alpha-1-antichymotrypsin | | Authors: | Schmidt, K, Muller, Y.A. | | Deposit date: | 2018-08-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.787 Å) | | Cite: | NewBG: A surrogate corticosteroid-binding globulin with an unprecedentedly high ligand release efficacy.
J.Struct.Biol., 207, 2019
|
|
6HGM
 
 | |
6KGA
 
 | |
2GD4
 
 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
3KCG
 
 | | Crystal structure of the antithrombin-factor IXa-pentasaccharide complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, Antithrombin-III, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of factor IXa recognition by heparin-activated antithrombin revealed by a 1.7-A structure of the ternary complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ZLZ
 
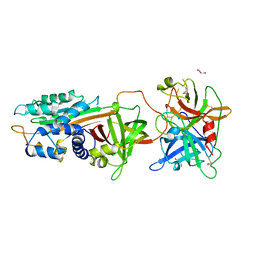 | | Structure of tPA and PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, Tissue-type plasminogen activator | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-03-31 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Development of a PAI-1 trapping agent (PAItrap2) based on inactivated tPA-SPD and the crystal structure of PAItrap2 in complex with PAI-1
To Be Published
|
|
8W18
 
 | |
7BEE
 
 | |
7BDU
 
 | |
5BRR
 
 | | Michaelis complex of tPA-S195A:PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, TRIETHYLENE GLYCOL, ... | | Authors: | Gong, L. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-02 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Crystal Structure of the Michaelis Complex between Tissue-type Plasminogen Activator and Plasminogen Activators Inhibitor-1
J.Biol.Chem., 290, 2015
|
|
3B9F
 
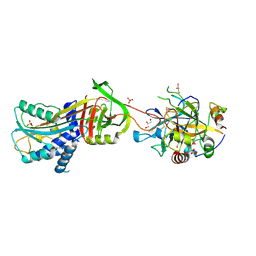 | | 1.6 A structure of the PCI-thrombin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, Plasma serine protease inhibitor, ... | | Authors: | Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7AKV
 
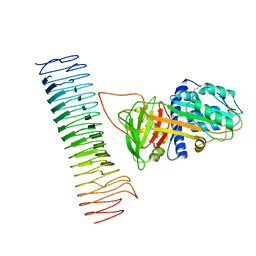 | | The cryo-EM structure of the Vag8-C1 inhibitor complex | | Descriptor: | Plasma protease C1 inhibitor, Vag8 | | Authors: | Johnson, S, Lea, S.M, Deme, J.C, Furlong, E, Dhillon, A. | | Deposit date: | 2020-10-02 | | Release date: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular Basis for Bordetella pertussis Interference with Complement, Coagulation, Fibrinolytic, and Contact Activation Systems: the Cryo-EM Structure of the Vag8-C1 Inhibitor Complex.
Mbio, 12, 2021
|
|
1OPH
 
 | | NON-COVALENT COMPLEX BETWEEN ALPHA-1-PI-PITTSBURGH AND S195A TRYPSIN | | Descriptor: | Alpha-1-antitrypsin precursor, Trypsinogen, cationic precursor | | Authors: | Dementiev, A, Simonovic, M, Volz, K, Gettins, P.G. | | Deposit date: | 2003-03-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Canonical inhibitor-like interactions explain reactivity of alpha1-proteinase inhibitor Pittsburgh and antithrombin with proteinases
J.Biol.Chem., 278, 2003
|
|
1OC0
 
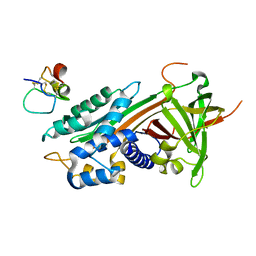 | | plasminogen activator inhibitor-1 complex with somatomedin B domain of vitronectin | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1, VITRONECTIN | | Authors: | Read, R.J, Zhou, A, Huntington, J.A, Pannu, N.S, Carrell, R.W. | | Deposit date: | 2003-02-03 | | Release date: | 2003-06-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | How Vitronectin Binds Pai-1 to Modulate Fibrinolysis and Cell Migration
Nat.Struct.Biol., 10, 2003
|
|
1SR5
 
 | | ANTITHROMBIN-ANHYDROTHROMBIN-HEPARIN TERNARY COMPLEX STRUCTURE | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2,3,6-tri-O-methyl-alpha-D-xylo-hexopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6S)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6R)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-1,5-anhydro-3-O-methyl-2,6-di-O-sulfo-D-glucitol, ... | | Authors: | Dementiev, A, Petitou, M, Gettins, P.G. | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2D26
 
 | |
1TB6
 
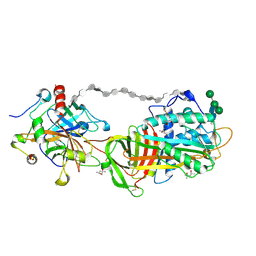 | | 2.5A Crystal Structure of the Antithrombin-Thrombin-Heparin Ternary Complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, W, Johnson, D.J, Esmon, C.T, Huntington, J.A. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the antithrombin-thrombin-heparin ternary complex reveals the antithrombotic mechanism of heparin.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2B5T
 
 | | 2.1 Angstrom structure of a nonproductive complex between antithrombin, synthetic heparin mimetic SR123781 and two S195A thrombin molecules | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-sulfonato-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-alpha-D-glucopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfonato-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-alpha-L-idopyranuronic acid-(1-4)-methyl 3-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Johnson, D.J, Li, W, Luis, S.A, Carrell, R.W, Huntington, J.A. | | Deposit date: | 2005-09-29 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of monomeric native antithrombin reveals a novel reactive center loop conformation.
J.Biol.Chem., 281, 2006
|
|
3PB1
 
 | | Crystal Structure of a Michaelis Complex between Plasminogen Activator Inhibitor-1 and Urokinase-type Plasminogen Activator | | Descriptor: | Plasminogen activator inhibitor 1, Plasminogen activator, urokinase, ... | | Authors: | Lin, Z, Jiang, L, Huang, M, Structure 2 Function Project (S2F) | | Deposit date: | 2010-10-20 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1.
J.Biol.Chem., 286, 2011
|
|
4DY7
 
 | |
3F1S
 
 | | Crystal structure of Protein Z complexed with protein Z-dependent inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, A. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of protein Z-dependent inhibitor complex shows how protein Z functions as a cofactor in the membrane inhibition of factor X.
Blood, 2009
|
|
