5ZLZ
 
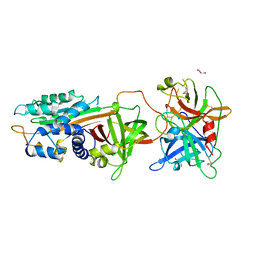 | | Structure of tPA and PAI-1 | | Descriptor: | GLYCEROL, Plasminogen activator inhibitor 1, Tissue-type plasminogen activator | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-03-31 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.581 Å) | | Cite: | Development of a PAI-1 trapping agent (PAItrap2) based on inactivated tPA-SPD and the crystal structure of PAItrap2 in complex with PAI-1
To Be Published
|
|
6AEX
 
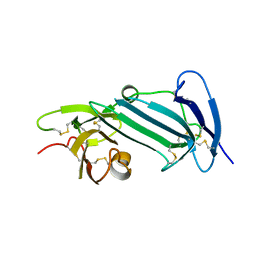 | | Crystal structure of unoccupied murine uPAR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Urokinase plasminogen activator surface receptor | | Authors: | Min, L, Huang, M. | | Deposit date: | 2018-08-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Crystal structure of the unoccupied murine urokinase-type plasminogen activator receptor (uPAR) reveals a tightly packed DII-DIII unit.
Febs Lett., 593, 2019
|
|
4H6Q
 
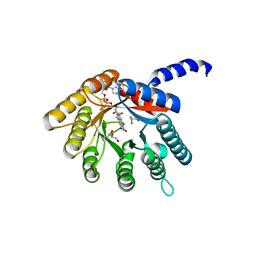 | |
4H6R
 
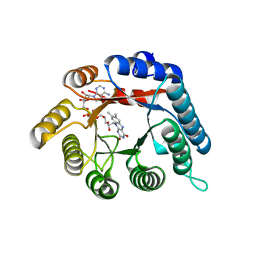 | | Structure of reduced Deinococcus radiodurans proline dehydrogenase | | Descriptor: | ACETATE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Proline dehydrogenase | | Authors: | Min, L, Tanner, J.J. | | Deposit date: | 2012-09-19 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures and kinetics of monofunctional proline dehydrogenase provide insight into substrate recognition and conformational changes associated with flavin reduction and product release.
Biochemistry, 51, 2012
|
|
3D2M
 
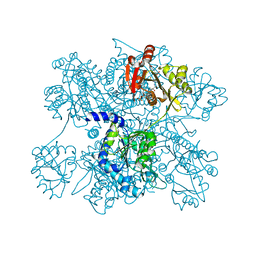 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-glutamate | | Descriptor: | COENZYME A, GLUTAMIC ACID, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
3D2P
 
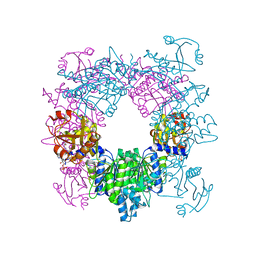 | | Crystal structure of N-acetylglutamate synthase from Neisseria gonorrhoeae complexed with coenzyme A and L-arginine | | Descriptor: | ARGININE, COENZYME A, Putative acetylglutamate synthase | | Authors: | Shi, D, Min, L, Jin, Z, Allewell, N.M, Tuchman, M. | | Deposit date: | 2008-05-08 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanism of Allosteric Inhibition of N-Acetyl-L-glutamate Synthase by L-Arginine.
J.Biol.Chem., 284, 2009
|
|
8HF4
 
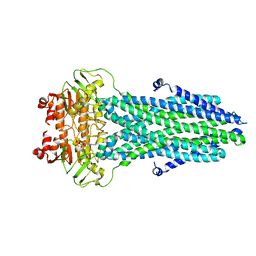 | |
8HF5
 
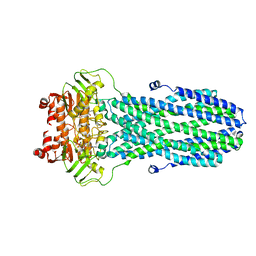 | |
8HF6
 
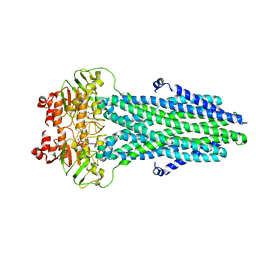 | | Cryo-EM structure of nucleotide-bound ComA E647Q mutant | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA | | Authors: | Yu, L, Xin, X, Min, L. | | Deposit date: | 2022-11-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA.
Nat Commun, 14, 2023
|
|
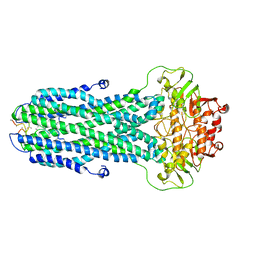
8HF7
 
 | |
8K4B
 
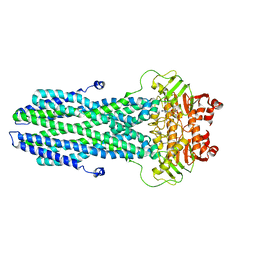 | | Cryo-EM structure of nucleotide-bound ComA with ZinC ion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Competence factor transporting ATP-binding protein/permease ComA, ZINC ION | | Authors: | Yu, L, Xin, X, Min, L, Feng, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-10-11 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of peptide secretion for Quorum sensing by ComA
Nat Commun, 14, 2023
|
|
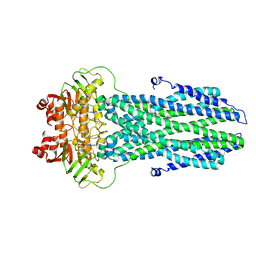
8K7A
 
 | |
8I6Q
 
 | |
