1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
2N3X
 
 | |
2N4H
 
 | |
2N4G
 
 | |
5GT1
 
 | | crystal structure of cbpa from L. salivarius REN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Choline binding protein A, ... | | Authors: | Jiang, L, Ren, F. | | Deposit date: | 2016-08-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins
Sci Rep, 7, 2017
|
|
3BBX
 
 | |
7VUE
 
 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
3KID
 
 | | The Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator | | Descriptor: | Urokinase-type plasminogen activator, ethyl 2-amino-1,3-benzothiazole-6-carboxylate | | Authors: | Jiang, L.-G, Yu, H.Y, Yuan, C, Wang, J.D, Chen, L.Q, Meehan, E.J, Huang, Z.-X, Huang, M.-D. | | Deposit date: | 2009-11-01 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
8HBM
 
 | |
4DVB
 
 | | The crystal structure of the Fab fragment of pro-uPA antibody mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
1TOB
 
 | | SACCHARIDE-RNA RECOGNITION IN AN AMINOGLYCOSIDE ANTIBIOTIC-RNA APTAMER COMPLEX, NMR, 7 STRUCTURES | | Descriptor: | 1,3-DIAMINO-5,6-DIHYDROXYCYCLOHEXANE, 2,6-diammonio-2,3,6-trideoxy-alpha-D-glucopyranose, 3-ammonio-3-deoxy-alpha-D-glucopyranose, ... | | Authors: | Jiang, L, Suri, A.K, Fiala, R, Patel, D.J. | | Deposit date: | 1996-12-12 | | Release date: | 1997-06-16 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in an aminoglycoside antibiotic-RNA aptamer complex.
Chem.Biol., 4, 1997
|
|
4DW2
 
 | | The crystal structure of uPA in complex with the Fab fragment of mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
4DVA
 
 | | The crystal structure of human urokinase-type plasminogen activator catalytic domain | | Descriptor: | HEXAETHYLENE GLYCOL, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody
Biochem.J., 449, 2013
|
|
3M61
 
 | | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition | | Descriptor: | Urokinase-type plasminogen activator, upain-1 W3A | | Authors: | Jiang, L, Yuan, C, Wind, T, Andreasen, P.A, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of complex of urokinase and a upain-1 variant(W3A) in pH4.6 condition
TO BE PUBLISHED
|
|
3MHW
 
 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
3BBU
 
 | |
4X1N
 
 | | The crystal structure of mupain-1-16 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X1Q
 
 | | The crystal structure of mupain-1 in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X0W
 
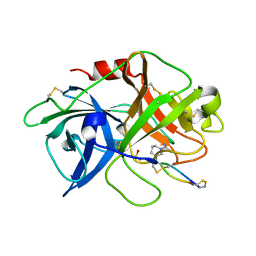 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1S
 
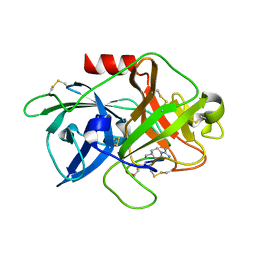 | | The crystal structure of mupain-1-16-D9A in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4X1R
 
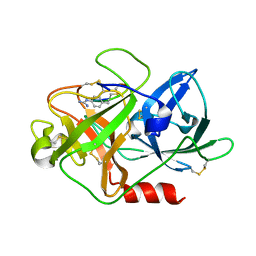 | | The crystal structure of mupain-1-12 in complex with murinised human uPA at pH7.4 | | Descriptor: | 1-phenylguanidine, Urokinase-type plasminogen activator, mupain-1-12 | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
4ZHM
 
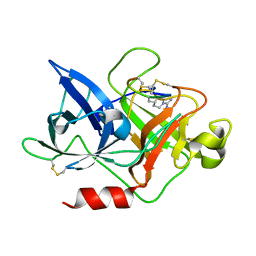 | |
4ZKO
 
 | |
4ZKR
 
 | |
