7UZX
 
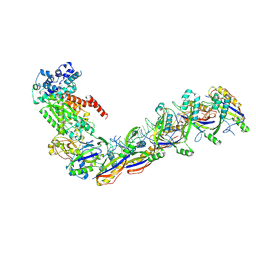 | | Staphylococcus epidermidis RP62a CRISPR effector subcomplex with non-self target RNA bound | | Descriptor: | CRISPR non-self RNA target, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm4, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
5DCA
 
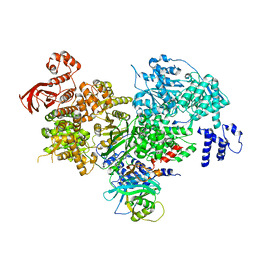 | | Crystal structure of yeast full length Brr2 in complex with Prp8 Jab1 domain | | Descriptor: | Pre-mRNA-splicing factor 8, Pre-mRNA-splicing helicase BRR2 | | Authors: | Absmeier, E, Wollenhaupt, J, Santos, K.F, Wahl, M.C. | | Deposit date: | 2015-08-23 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation.
Genes Dev., 29, 2015
|
|
7V00
 
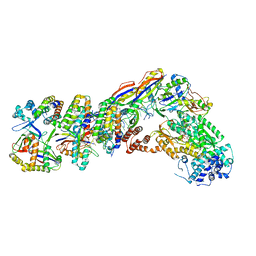 | | Staphylococcus epidermidis RP62a CRISPR tall effector complex with bound ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, ... | | Authors: | Smith, E.M, Ferrell, S.H, Tokars, V.L, Mondragon, A. | | Deposit date: | 2022-05-09 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Structures of an active type III-A CRISPR effector complex.
Structure, 30, 2022
|
|
7M71
 
 | | SARS-CoV-2 Spike:5A6 Fab complex I focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 5A6 Fab heavy chain, Antibody 5A6 Fab light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
7M7B
 
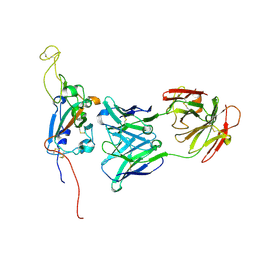 | | SARS-CoV-2 Spike:Fab 3D11 complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 3D11 heavy chain, Antibody Fab 3D11 light chain, ... | | Authors: | Asarnow, D, Cheng, Y. | | Deposit date: | 2021-03-27 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia.
Cell, 184, 2021
|
|
8JPN
 
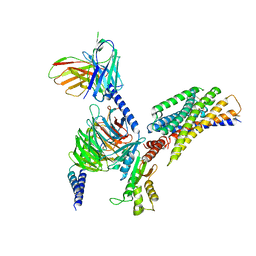 | | Cryo-EM structure of succinate receptor bound to cis-epoxysuccinic acid coupling to Gi | | Descriptor: | (2R,3S)-oxirane-2,3-dicarboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, T.X, Tang, W.Q, Li, F.H, Wang, J.Y. | | Deposit date: | 2023-06-12 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular activation and G protein coupling selectivity of human succinate receptor SUCR1.
Cell Res., 34, 2024
|
|
6S2O
 
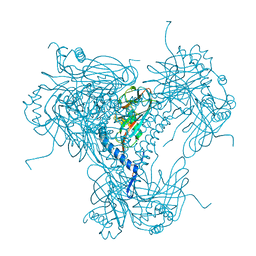 | |
7UM7
 
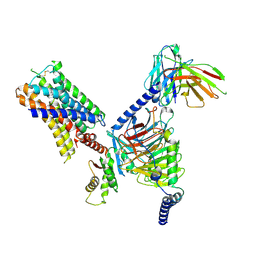 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Methylergometrine | | Descriptor: | (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UFG
 
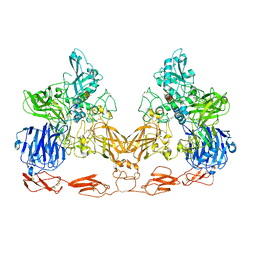 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
7UM5
 
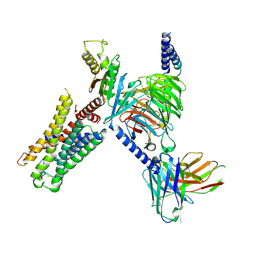 | | CryoEM structure of Go-coupled 5-HT5AR in complex with 5-CT | | Descriptor: | 3-(2-azanylethyl)-1H-indole-5-carboxamide, 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8IRT
 
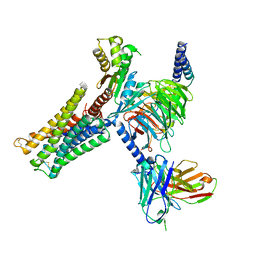 | | Dopamine Receptor D3R-Gi-Rotigotine complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
8IRR
 
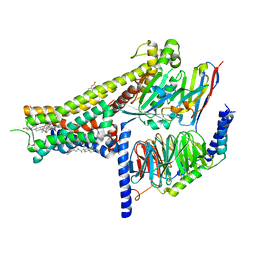 | | Dopamine Receptor D1R-Gs-Rotigotine complex | | Descriptor: | CHOLESTEROL, D(1A) dopamine receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, P, Huang, S, Zhuang, Y, Mao, C, Zhang, Y, Wang, Y, Li, H, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2023-03-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural genomics of the human dopamine receptor system.
Cell Res., 33, 2023
|
|
7UM6
 
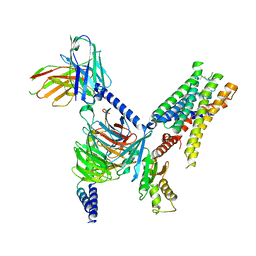 | | CryoEM structure of Go-coupled 5-HT5AR in complex with Lisuride | | Descriptor: | 5-hydroxytryptamine receptor 5A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhang, S, Fay, J.F, Roth, B.L. | | Deposit date: | 2022-04-06 | | Release date: | 2022-07-20 | | Last modified: | 2022-07-27 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Inactive and active state structures template selective tools for the human 5-HT 5A receptor.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DP0
 
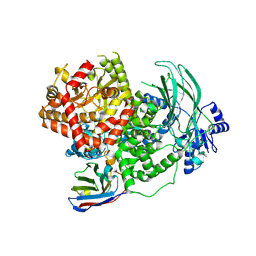 | | Structure of p110 gamma bound to the Ras inhibitory nanobody NB7 | | Descriptor: | Nanobody NB7, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Burke, J.E, Nam, S.E, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Allosteric activation or inhibition of PI3K gamma mediated through conformational changes in the p110 gamma helical domain.
Elife, 12, 2023
|
|
8J6J
 
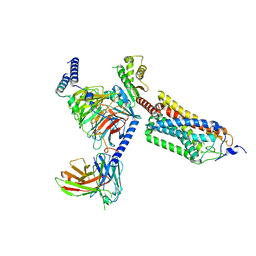 | | Cryo-EM structure of thehydroxycarboxylic acid receptor 2-Gi protein complex bound with GSK256073 | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yuan, Q, Zhu, S, Duan, J, Xu, H.E, Duan, X. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Cell Rep, 42, 2023
|
|
8JGG
 
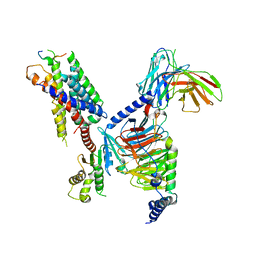 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist BAM8-22 | | Descriptor: | BAM8-22, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Ynag, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8JGB
 
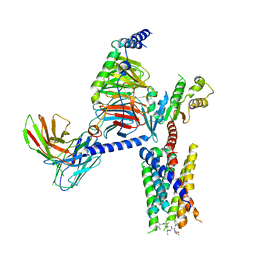 | | CryoEM structure of Gi-coupled MRGPRX1 with peptide agonist CNF-Tx2 | | Descriptor: | Conorfamide-Tx2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Xu, H.E, Yang, F, Liu, Z.M, Guo, L.L, Zhang, Y.M, Fang, G.X, Tie, L, Zhuang, Y.M, Xue, C.Y. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G protein coupling of the human itch receptor MRGPRX1.
Nat Commun, 14, 2023
|
|
8J22
 
 | | Cryo-EM structure of FFAR2 complex bound with TUG-1375 | | Descriptor: | (2R,4R)-2-(2-chlorophenyl)-3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]carbonyl-1,3-thiazolidine-4-carboxylic acid, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J20
 
 | | Cryo-EM structure of FFAR3 bound with valeric acid and AR420626 | | Descriptor: | (4R)-N-[2,5-bis(chloranyl)phenyl]-4-(furan-2-yl)-2-methyl-5-oxidanylidene-4,6,7,8-tetrahydro-1H-quinoline-3-carboxamide, Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
8J24
 
 | | Cryo-EM structure of FFAR2 complex bound with acetic acid | | Descriptor: | ACETATE ION, Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Tai, L, Li, F, Tang, W, Sun, X, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
6K4M
 
 | |
8J21
 
 | | Cryo-EM structure of FFAR3 complex bound with butyrate acid | | Descriptor: | Free fatty acid receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular recognition and activation mechanism of short-chain fatty acid receptors FFAR2/3.
Cell Res., 34, 2024
|
|
6HUG
 
 | | CryoEM structure of human full-length alpha1beta3gamma2L GABA(A)R in complex with picrotoxin and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
6HUO
 
 | | CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with alprazolam (Xanax), GABA and megabody Mb38. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 8-chloro-1-methyl-6-phenyl-4H-[1,2,4]triazolo[4,3-a][1,4]benzodiazepine, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-09 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
6HUJ
 
 | | CryoEM structure of human full-length heteromeric alpha1beta3gamma2L GABA(A)R in complex with picrotoxin, GABA and megabody Mb38. | | Descriptor: | (1aR,2aR,3S,6R,6aS,8aS,8bR,9R)-2a-hydroxy-8b-methyl-9-(prop-1-en-2-yl)hexahydro-3,6-methano-1,5,7-trioxacyclopenta[ij]c yclopropa[a]azulene-4,8(3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Masiulis, S, Desai, R, Uchanski, T, Serna Martin, I, Laverty, D, Karia, D, Malinauskas, T, Jasenko, Z, Pardon, E, Kotecha, A, Steyaert, J, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2018-10-08 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | GABAAreceptor signalling mechanisms revealed by structural pharmacology.
Nature, 565, 2019
|
|
