5KQR
 
 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine | | Descriptor: | CHLORIDE ION, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Jain, R, Coloma, J, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
2ZMB
 
 | | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with parecoxib at 2.9 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Jain, R, Mir, R, Sinha, M, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-15 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the complex of C-terminal lobe of bovine lactoferrin with parecoxib at 2.9 A resolution
To be Published
|
|
3EZX
 
 | |
1XEO
 
 | | High Resolution Crystals Structure of Cobalt- Peptide Deformylase Bound To Formate | | Descriptor: | COBALT (II) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEM
 
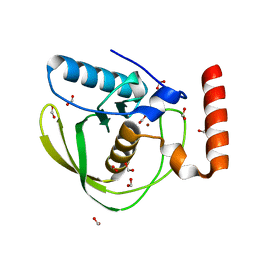 | | High Resolution Crystal Structure of Escherichia coli Zinc- Peptide Deformylase bound to formate | | Descriptor: | FORMIC ACID, Peptide deformylase, ZINC ION | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
1XEN
 
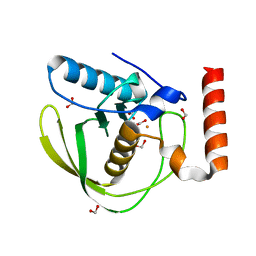 | | High Resolution Crystal Structure of Escherichia coli Iron- Peptide Deformylase Bound To Formate | | Descriptor: | FE (III) ION, FORMIC ACID, Peptide deformylase | | Authors: | Jain, R, Hao, B, Liu, R.-P, Chan, M.K. | | Deposit date: | 2004-09-10 | | Release date: | 2005-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of E. coli peptide deformylase bound to formate: insight into the preference for Fe2+ over Zn2+ as the active site metal
J.Am.Chem.Soc., 127, 2005
|
|
4PTF
 
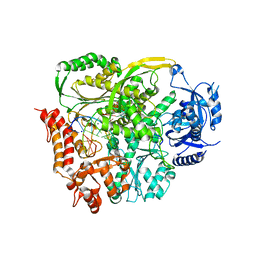 | | Ternary crystal structure of yeast DNA polymerase epsilon with template G | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*AP*TP*CP*CP*TP*CP*CP*CP*CP*TP*AP*(DOC))-3', ... | | Authors: | Jain, R, Rajashankar, K.R, Buku, A, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2014-03-10 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Crystal Structure of Yeast DNA Polymerase epsilon Catalytic Domain.
Plos One, 9, 2014
|
|
3H4B
 
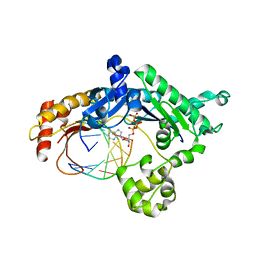 | | Ternary complex of human DNA polymerase iota with template U/T and incoming dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(*TP*(BRU)P*GP*GP*GP*TP*CP*CP*T)-3', ... | | Authors: | Jain, R, Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2009-04-18 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Replication across template T/U by human DNA polymerase-iota.
Structure, 17, 2009
|
|
3H40
 
 | | Binary complex of human DNA polymerase iota with template U/T | | Descriptor: | 5'-D(*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(*TP*(BRU)P*GP*GP*GP*TP*CP*CP*T)-3', DNA polymerase iota, ... | | Authors: | Jain, R, Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Replication across template T/U by human DNA polymerase-iota.
Structure, 17, 2009
|
|
3H4D
 
 | | Ternary complex of human DNA polymerase iota with template U/T and incoming dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*AP*CP*CP*(DOC)), 5'-D(*TP*(BRU)P*GP*GP*GP*TP*CP*CP*T), ... | | Authors: | Jain, R, Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2009-04-18 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Replication across template T/U by human DNA polymerase-iota.
Structure, 17, 2009
|
|
5ULX
 
 | |
5ULW
 
 | |
1Q4B
 
 | |
1Q4E
 
 | |
1Q4A
 
 | |
1Q4D
 
 | |
1Q4C
 
 | |
1Q73
 
 | |
6P1H
 
 | | Cryo-EM Structure of DNA Polymerase Delta Holoenzyme | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Jain, R, Rice, W, Aggarwal, A.K. | | Deposit date: | 2019-05-19 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure and dynamics of eukaryotic DNA polymerase delta holoenzyme.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5ULP
 
 | | Structure of the NS5 methyltransferase from Zika bound to MS2042 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][(4-fluorophenyl)methyl]amino}-5'-deoxyadenosine, CHLORIDE ION, ISOPROPYL ALCOHOL, ... | | Authors: | Jain, R, Aggarwal, A.K. | | Deposit date: | 2017-01-25 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of a S-adenosylmethionine analog that intrudes the RNA-cap binding site of Zika methyltransferase.
Sci Rep, 7, 2017
|
|
5JRZ
 
 | |
2PWA
 
 | | Crystal Structure of the complex of Proteinase K with Alanine Boronic acid at 0.83A resolution | | Descriptor: | ALANINE BORONIC ACID, CALCIUM ION, NITRATE ION, ... | | Authors: | Jain, R, Singh, N, Perbandt, M, Betzel, C, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Crystal structure of the complex of Proteinase K with Alanine Boronic Acid at 0.83A Resolution
To be Published
|
|
4EEY
 
 | | Crystal structure of human DNA polymerase eta in ternary complex with a cisplatin DNA adduct | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*TP*TP*GP*GP*TP*CP*TP*CP*CP*TP*CP*C)-3', 5'-D(*TP*GP*GP*AP*GP*GP*AP*GP*A)-3', ... | | Authors: | Ummat, A, Rechkoblit, O, Jain, R, Choudhury, J.R, Johnson, R.E, Silverstein, T.D, Buku, A, Lone, S, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for cisplatin DNA damage tolerance by human polymerase {eta} during cancer chemotherapy.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5KQS
 
 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine and 7-methyl-guanosine-5'-diphosphate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, ACETATE ION, GLYCEROL, ... | | Authors: | Coloma, J, Jain, R, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
2Z5Z
 
 | | Crystal structure of the complex of buffalo Lactoperoxidase with fluoride ion at 3.5A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sheikh, I.A, Jain, R, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the complex of buffalo Lactoperoxidase with fluoride ion at 3.5A resolution
To be Published
|
|
