7T5G
 
 | | Structure of rabies virus phosphoprotein C-terminal domain, S210E mutant | | Descriptor: | Phosphoprotein, SULFATE ION | | Authors: | Zhan, J, Metcalfe, R.D, Gooley, P.R, Griffin, M.D.W. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of Functional Effects of Phosphorylation of the C-Terminal Domain of the Rabies Virus P Protein.
J.Virol., 96, 2022
|
|
2HDC
 
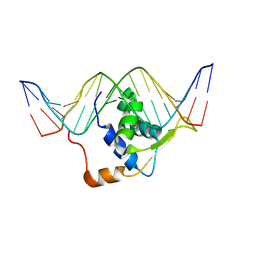 | | STRUCTURE OF TRANSCRIPTION FACTOR GENESIS/DNA COMPLEX | | Descriptor: | DNA (5'-D(P*GP*CP*TP*TP*AP*AP*AP*AP*TP*AP*AP*CP*AP*AP*TP*AP*C)-3'), DNA (5'-D(P*GP*TP*AP*TP*TP*GP*TP*TP*AP*TP*TP*TP*TP*AP*AP*GP*C)-3'), PROTEIN (TRANSCRIPTION FACTOR) | | Authors: | Jin, C, Marsden, I, Chen, X, Liao, X. | | Deposit date: | 1999-05-05 | | Release date: | 1999-07-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex.
J.Mol.Biol., 289, 1999
|
|
2HFH
 
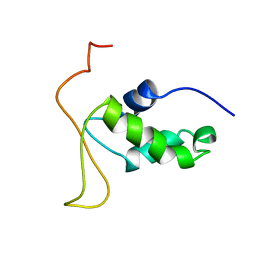 | |
4QP1
 
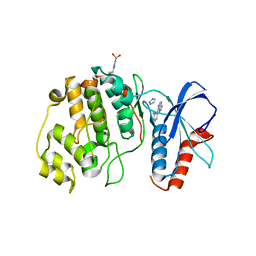 | |
4QP9
 
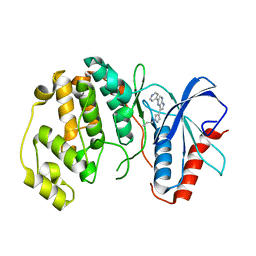 | |
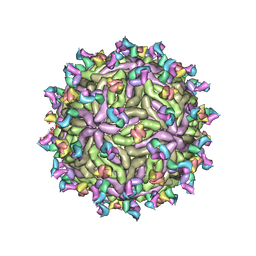
7T17
 
 | |
4QP3
 
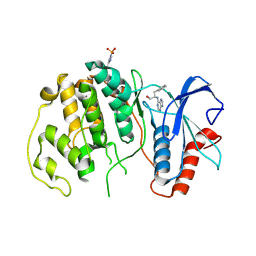 | |
8PT5
 
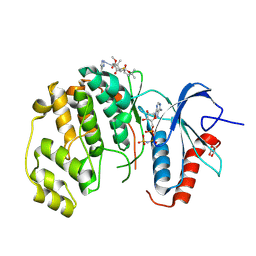 | | ERK2 covelently bound to RU187 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
4QP6
 
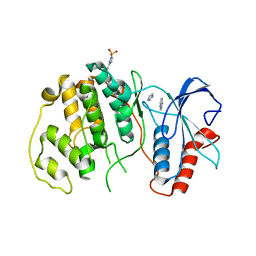 | |
8PSY
 
 | | ERK2 covelently bound to RU68 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{S},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PSW
 
 | | ERK2 covalently bound to RU67 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{R})-4-oxidanylidene-1-(phenylmethyl)cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT1
 
 | | ERK2 covelently bound to RU76 cyclohexenone based inhibitor | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ~{O}1-methyl ~{O}3-(phenylmethyl) (1~{S},3~{R})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT3
 
 | | ERK2 covelently bound to RU77 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PST
 
 | | ERK2 covelently bound to RU60 cyclohexenone based inhibitor | | Descriptor: | AMP PHOSPHORAMIDATE, Mitogen-activated protein kinase 1, ~{O}3-~{tert}-butyl ~{O}1-methyl (1~{R},3~{S})-1-methyl-4-oxidanylidene-cyclohexane-1,3-dicarboxylate | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
3IH7
 
 | |
8PSR
 
 | | ERK2 covalently bound to SynthRevD-12-opt artificial peptide | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Gogl, G, Remenyi, A. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
8PT0
 
 | | ERK2 covelently bound to RU75 cyclohexenone based inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Sok, P, Poti, A, Remenyi, A, Gogl, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a Michael acceptor-based cyclic warhead scaffold
To Be Published
|
|
5SYA
 
 | |
3WYG
 
 | | Crystal structure of Xpo1p-PKI-Gsp1p-GTP complex | | Descriptor: | Exportin-1, GUANOSINE-5'-TRIPHOSPHATE, Gsp1p, ... | | Authors: | Koyama, M, Shirai, N, Matsuura, Y. | | Deposit date: | 2014-08-26 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into how yrb2p accelerates the assembly of the xpo1p nuclear export complex
Cell Rep, 9, 2014
|
|
8GO8
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C5a anaphylatoxin chemotactic receptor 1, C5aR1 | | Descriptor: | Beta-arrestin-1, C5a anaphylatoxin chemotactic receptor 1, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8GP3
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
4QTE
 
 | | Structure of ERK2 in complex with VTX-11e, 4-{2-[(2-CHLORO-4-FLUOROPHENYL)AMINO]-5-METHYLPYRIMIDIN-4-YL}-N-[(1S)-1-(3-CHLOROPHENYL)-2-HYDROXYETHYL]-1H-PYRROLE-2-CARBOXAMIDE | | Descriptor: | 1,2-ETHANEDIOL, 4-{2-[(2-chloro-4-fluorophenyl)amino]-5-methylpyrimidin-4-yl}-N-[(1S)-1-(3-chlorophenyl)-2-hydroxyethyl]-1H-pyrrole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Chaikuad, A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-07 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics.
Nat.Chem.Biol., 10, 2014
|
|
5NPH
 
 | |
5I5Z
 
 | | CDK8-CYCC IN COMPLEX WITH 8-(1-Methyl-2,2-dioxo-2,3-dihydro-1H-2l6-benzo[c]isothiazol-5-yl)-[1,6]naphthyridine-2-carboxylic acid methylamide | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, FORMIC ACID, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A. | | Deposit date: | 2016-02-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2,8-Disubstituted-1,6-Naphthyridines and 4,6-Disubstituted-Isoquinolines with Potent, Selective Affinity for CDK8/19.
Acs Med.Chem.Lett., 7, 2016
|
|
2IC3
 
 | |
