6P95
 
 | | Structure of Lassa virus glycoprotein in complex with Fab 25.6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Hastie, K.M. | | Deposit date: | 2019-06-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Convergent Structures Illuminate Features for Germline Antibody Binding and Pan-Lassa Virus Neutralization.
Cell, 178, 2019
|
|
6PCI
 
 | | EBOV GPdMuc (Makona) in complex with rEBOV-520 and rEBOV-548 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Virion spike glycoprotein, Virion spike glycoprotein,Virion spike glycoprotein,Ebola Virus (Makona) GP2, ... | | Authors: | Ward, A.B, Murin, C.D, Alkutkar, T. | | Deposit date: | 2019-06-17 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
6PV7
 
 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with nicotine | | Descriptor: | (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
6PV8
 
 | | Human alpha3beta4 nicotinic acetylcholine receptor in complex with AT-1001 | | Descriptor: | (3-endo)-N-(2-bromophenyl)-9-methyl-9-azabicyclo[3.3.1]nonan-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gharpure, A, Teng, J, Zhuang, Y, Noviello, C.M, Walsh, R.M, Cabuco, R, Howard, R.J, Zaveri, N.T, Lindahl, E, Hibbs, R.E. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Agonist Selectivity and Ion Permeation in the alpha 3 beta 4 Ganglionic Nicotinic Receptor.
Neuron, 104, 2019
|
|
6PWU
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-07-23 | | Release date: | 2020-02-26 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6PXH
 
 | | Crystal Structure of MERS-CoV S1-NTD bound with G2 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DIHYDROFOLIC ACID, ... | | Authors: | Wang, N, McLellan, J.S. | | Deposit date: | 2019-07-26 | | Release date: | 2019-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6PZ8
 
 | | MERS S0 trimer in complex with variable domain of antibody G2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G2 heavy chain, ... | | Authors: | Bowman, C.A, Pallesen, J, Ward, A.B. | | Deposit date: | 2019-07-31 | | Release date: | 2019-10-09 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural Definition of a Neutralization-Sensitive Epitope on the MERS-CoV S1-NTD.
Cell Rep, 28, 2019
|
|
6Q04
 
 | | MERS-CoV S structure in complex with 5-N-acetyl neuraminic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q05
 
 | | MERS-CoV S structure in complex with sialyl-lewisX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q06
 
 | | MERS-CoV S structure in complex with 2,3-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6Q07
 
 | | MERS-CoV S structure in complex with 2,6-sialyl-N-acetyl-lactosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FOLIC ACID, ... | | Authors: | Park, Y.J, Walls, A.C, Wang, Z, Sauer, M, Li, W, Tortorici, M.A, Bosch, B.J, DiMaio, F.D, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-01 | | Release date: | 2019-12-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6RU5
 
 | | human complement C3 in complex with the hC3Nb1 nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2019-05-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
6S1X
 
 | |
6S9J
 
 | | Crystal structure of TfR1 mimicry in complex with GP1 from MACV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pre-glycoprotein polyprotein GP complex, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-07-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Rational design of universal immunotherapy for TfR1-tropic arenaviruses.
Nat Commun, 11, 2020
|
|
6SGP
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a sulfamide inhibitor GluGlu | | Descriptor: | (2~{S})-2-[[(2~{S})-1,5-bis(oxidanyl)-1,5-bis(oxidanylidene)pentan-2-yl]sulfamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barinka, C, Shukla, S, Motlova, L. | | Deposit date: | 2019-08-05 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
6SKH
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) - the E424M inactive mutant, in complex with a inhibitor sulfamide inhibitor GluAsp | | Descriptor: | (2~{S})-2-[[(2~{S})-1,4-bis(oxidanyl)-1,4-bis(oxidanylidene)butan-2-yl]sulfamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2019-08-15 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural, Biochemical, and Computational Characterization of Sulfamides as Bimetallic Peptidase Inhibitors.
J.Chem.Inf.Model., 2024
|
|
6TS2
 
 | | Truncated version of Chaetomium thermophilum UDP-Glucose Glucosyl Transferase (UGGT) lacking domain TRXL2 (417-650). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein,UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N. | | Deposit date: | 2019-12-19 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (5.74 Å) | | Cite: | Clamping, bending, and twisting inter-domain motions in the misfold-recognizing portion of UDP-glucose: Glycoprotein glucosyltransferase.
Structure, 29, 2021
|
|
6TVA
 
 | | Crystal structure of the haemagglutinin from a transmissible H10N7 seal influenza virus isolated in Netherland in complex with avian receptor analogue, 3'-SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Haemagglutinin HA1, Haemagglutinin HA2, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6TY5
 
 | | Crystal structure of human TLR8 in complex with Compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-methyl-7-(7-methyl-2-piperidin-4-yl-indazol-5-yl)furo[3,2-c]pyridin-4-one, Toll-like receptor 8, ... | | Authors: | Faller, M, Zink, F. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Target-Based Identification and Optimization of 5-Indazol-5-yl Pyridones as Toll-like Receptor 7 and 8 Antagonists Using a Biochemical TLR8 Antagonist Competition Assay.
J.Med.Chem., 63, 2020
|
|
6TYB
 
 | |
6UDJ
 
 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-04-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
6UQR
 
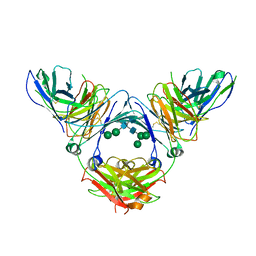 | | Complex of IgE and Ligelizumab | | Descriptor: | IgE, Ligelizumab, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tarchevskaya, S.S, Kleinboelting, S, Jardetzky, T.S. | | Deposit date: | 2019-10-21 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.65029109708 Å) | | Cite: | The mechanistic and functional profile of the therapeutic anti-IgE antibody ligelizumab differs from omalizumab.
Nat Commun, 11, 2020
|
|
6UTK
 
 | |
6UYE
 
 | | EBOV GPdMuc Makona bound to rEBOV-548 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody rEBOV-548 Fab, ... | | Authors: | Murin, C.D, Ward, A.B. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Analysis of a Therapeutic Antibody Cocktail Reveals Determinants for Cooperative and Broad Ebolavirus Neutralization.
Immunity, 52, 2020
|
|
