4CEW
 
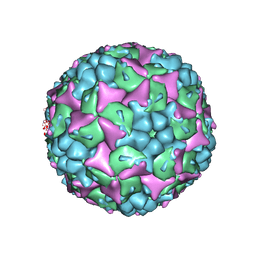 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3MUZ
 
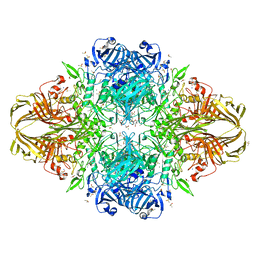 | | E.Coli (lacZ) beta-galactosidase (R599A) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M.L, Driedger, M.R, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
3ZLV
 
 | | Crystal structure of mouse acetylcholinesterase in complex with tabun and HI-6 | | Descriptor: | (2-hydroxyethoxy)acetaldehyde, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Artursson, E, Andersson, P.O, Akfur, C, Linusson, A, Borjegren, S, Ekstrom, F. | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic-Site Conformational Equilibrium in Nerve-Agent Adducts of Acetylcholinesterase; Possible Implications for the Hi-6 Antidote Substrate Specificity.
Biochem.Pharmacol., 85, 2013
|
|
4RJY
 
 | |
1Y6V
 
 | | Structure of E. coli Alkaline Phosphatase in presence of cobalt at 1.60 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
2PLK
 
 | |
3MV0
 
 | | E. COLI (lacZ) BETA-GALACTOSIDASE (R599A) IN COMPLEX WITH D-GALCTOPYRANOSYL-1-ONE | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Dugdale, M.L, Vance, M, Driedger, M.L, Nibber, A, Tran, A, Huber, R.E. | | Deposit date: | 2010-05-03 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of Arg-599 of b-galactosidase (Escherichia coli) as an anchor for the open conformations of Phe-601 and the active-site loop
Biochem.Cell Biol., 88, 2010
|
|
2ZTN
 
 | | Hepatitis E virus ORF2 (Genotype 3) | | Descriptor: | Capsid protein | | Authors: | Yamashita, T, Unno, H, Mori, Y, Li, T.C, Takeda, N, Matsuura, Y. | | Deposit date: | 2008-10-08 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5CYQ
 
 | | HIV-1 reverse transcriptase complexed with 4-bromopyrazole | | Descriptor: | 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, BROMIDE ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Rapid experimental SAD phasing and hot-spot identification with halogenated fragments.
IUCrJ, 3, 2016
|
|
1G1T
 
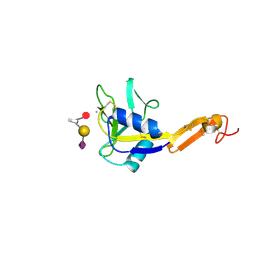 | | CRYSTAL STRUCTURE OF E-SELECTIN LECTIN/EGF DOMAINS COMPLEXED WITH SLEX | | Descriptor: | CALCIUM ION, E-SELECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Somers, W.S, Camphausen, R.T. | | Deposit date: | 2000-10-13 | | Release date: | 2001-10-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1.
Cell(Cambridge,Mass.), 103, 2000
|
|
4PPP
 
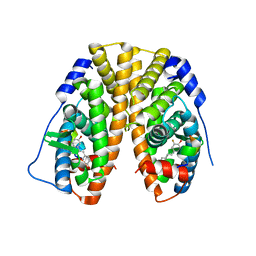 | | Crystal Structure of the Estrogen Receptor alpha Ligand-binding Domain in Complex with Fluoro-Resveratrol | | Descriptor: | 5-[(E)-2-(3-fluoro-4-hydroxyphenyl)ethenyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Parent, A.A, Hughes, T.S, Pollock, J.A, Gjyshi, O, Cavett, V, Nowak, J, Garcia-Ordonez, R.D, Houtman, R, Griffin, P.R, Kojetin, D.J, Katzenellenbogen, J.A, Conkright, M.D, Nettles, K.W. | | Deposit date: | 2014-02-27 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.686 Å) | | Cite: | Resveratrol modulates the inflammatory response via an estrogen receptor-signal integration network.
Elife, 3, 2014
|
|
2GNK
 
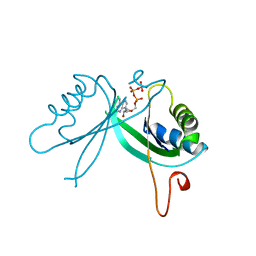 | | GLNK, A SIGNAL PROTEIN FROM E. COLI | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (NITROGEN REGULATORY PROTEIN) | | Authors: | Xu, Y, Cheah, E, Carr, P.D, van Heeswijk, W.C, Westerhoff, H.V, Vasudevan, S.G, Ollis, D.L. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GlnK, a PII-homologue: structure reveals ATP binding site and indicates how the T-loops may be involved in molecular recognition.
J.Mol.Biol., 282, 1998
|
|
3WYJ
 
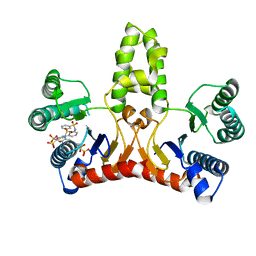 | | Structure of E. coli undecaprenyl diphosphate synthase in complex with BPH-789 | | Descriptor: | Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific), [1-oxidanyl-2-[3-[3-[[3-[[3-[3-(2-oxidanyl-2,2-diphosphono-ethyl)phenyl]phenyl]sulfamoyl]phenyl]sulfonylamino]phenyl]phenyl]-1-phosphono-ethyl]phosphonic acid | | Authors: | Gao, J, Ko, T.P, Huang, C.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-08-29 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
5DGS
 
 | |
3H1C
 
 | |
3S7K
 
 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
2FEQ
 
 | | orally active thrombin inhibitors | | Descriptor: | Decapeptide Hirudin Analogue, N-(CARBOXYMETHYL)-3-CYCLOHEXYL-D-ALANYL-N-({4-[(E)-AMINO(IMINO)METHYL]-1,3-THIAZOL-2-YL}METHYL)-L-PROLINAMIDE, Thrombin heavy chain, ... | | Authors: | Mack, H, Baucke, D, Hornberger, W, Lange, U.E.W, Hoeffken, H.W. | | Deposit date: | 2005-12-16 | | Release date: | 2006-08-08 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Orally active thrombin inhibitors. Part 1: optimization of the P1-moiety
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2LP8
 
 | | SOLUTION STRUCTURE OF AN APOPTOSIS ACTIVATING PHOTOSWITCHABLE BAK PEPTIDE BOUND to BCL-XL | | Descriptor: | 3,3'-(E)-diazene-1,2-diylbis{6-[(chloroacetyl)amino]benzenesulfonic acid}, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 1 | | Authors: | Wysoczanski, P, Mart, R.J, Loveridge, J.E, Williams, C, Whittaker, S.B.-M, Crump, M.P, Allemann, R.K. | | Deposit date: | 2012-02-03 | | Release date: | 2012-04-18 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Photoswitchable Apoptosis Activating Bak Peptide Bound to Bcl-x(L).
J.Am.Chem.Soc., 134, 2012
|
|
4DKP
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with AWS-I-50 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-amino-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7978 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
3AAU
 
 | | Bovine beta-trypsin bound to meta-diguanidino schiff base copper (II) chelate | | Descriptor: | CALCIUM ION, COPPER (II) ION, Cationic trypsin, ... | | Authors: | Iyaguchi, D, Kawano, S, Toyota, E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the design of novel Schiff base metal chelate inhibitors of trypsin
Bioorg.Med.Chem., 18, 2010
|
|
1TOG
 
 | | Hydrocinnamic acid-bound structure of SRHEPT + A293D mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, HYDROCINNAMIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
4QMO
 
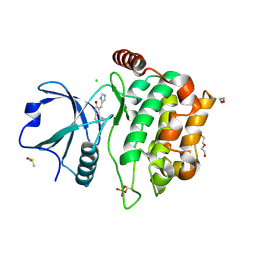 | | MST3 IN COMPLEX WITH Imidazolo-oxindole PKR inhibitor C16 | | Descriptor: | (8Z)-8-(1H-imidazol-5-ylmethylidene)-6,8-dihydro-7H-[1,3]thiazolo[5,4-e]indol-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
4DT0
 
 | |
4RDV
 
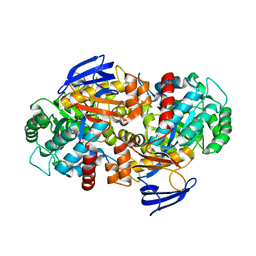 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
4MIT
 
 | |
