2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3RUK
 
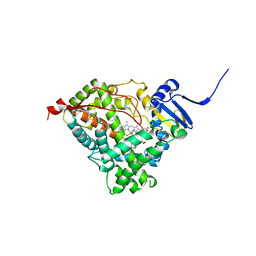 | | Human Cytochrome P450 CYP17A1 in complex with Abiraterone | | Descriptor: | Abiraterone, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of cytochrome P450 17A1 with prostate cancer drugs abiraterone and TOK-001.
Nature, 482, 2012
|
|
1Z23
 
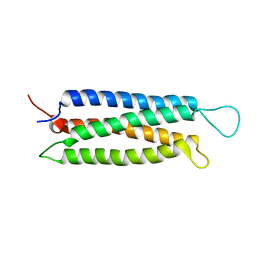 | | The serine-rich domain from Crk-associated substrate (p130Cas) | | Descriptor: | CRK-associated substrate | | Authors: | Briknarova, K, Nasertorabi, F, Havert, M.L, Eggleston, E, Hoyt, D.W, Li, C, Olson, A.J, Vuori, K, Ely, K.R. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The serine-rich domain from Crk-associated substrate (p130cas) is a four-helix bundle.
J.Biol.Chem., 280, 2005
|
|
2JEV
 
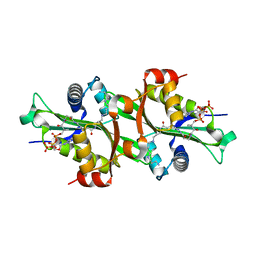 | | Crystal structure of human spermine,spermidine acetyltransferase in complex with a bisubstrate analog (N1-acetylspermine-S-CoA). | | Descriptor: | (3R)-27-AMINO-3-HYDROXY-2,2-DIMETHYL-4,8,14-TRIOXO-12-THIA-5,9,15,19,24-PENTAAZAHEPTACOS-1-YL [(2S,3R,4S,5S)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, DIAMINE ACETYLTRANSFERASE 1 | | Authors: | Hegde, S.S, Chandler, J, Vetting, M.W, Yu, M, Blanchard, J.S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic and Structural Analysis of Human Spermidine/Spermine N(1)-Acetyltransferase.
Biochemistry, 46, 2007
|
|
3K1E
 
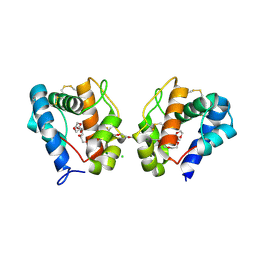 | | Crystal structure of odorant binding protein 1 (AaegOBP1) from Aedes aegypti | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Leite, N.R, Krogh, R, Leal, W.S, Iulek, J, Oliva, G. | | Deposit date: | 2009-09-27 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive "Lid".
Plos One, 4, 2009
|
|
3F69
 
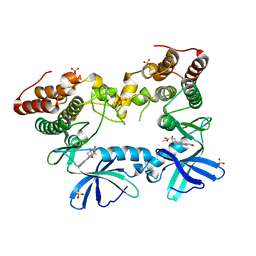 | | Crystal structure of the Mycobacterium tuberculosis PknB mutant kinase domain in complex with KT5720 | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase pknB, hexyl (5S,6R,8R)-6-hydroxy-5-methyl-13-oxo-5,6,7,8-tetrahydro-13H-5,8-epoxy-4b,8a,14-triazadibenzo[b,h]cycloocta[1,2,3,4-jkl]c yclopenta[e]-as-indacene-6-carboxylate | | Authors: | Alber, T, Mieczkowski, C.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase.
Embo J., 27, 2008
|
|
1MX9
 
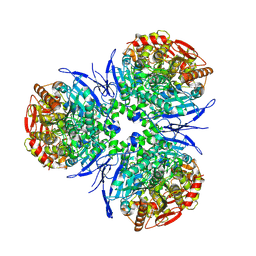 | | Crystal Structure of Human Liver Carboxylesterase in complexed with naloxone methiodide, a heroin analogue | | Descriptor: | (5A,17R)-4,5-EPOXY-3,14-DIHYDROXY-17-METHYL-6-OXO-17-(2-PROPENYL)-MORPHINANIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, liver Carboxylesterase I | | Authors: | Bencharit, S, Morton, C.L, Xue, Y, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2002-10-01 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Heroin and Cocaine Metabolism by a Promiscuous Human Drug-Processing Enzyme
Nat.Struct.Biol., 10, 2003
|
|
1NHZ
 
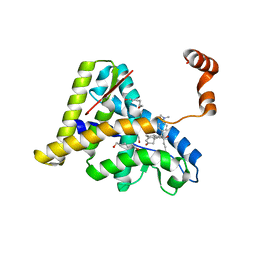 | | Crystal Structure of the Antagonist Form of Glucocorticoid Receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLUCOCORTICOID RECEPTOR, HEXANE-1,6-DIOL | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
4XVH
 
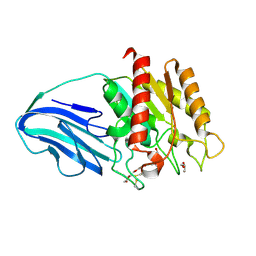 | | Crystal structure of a Corynascus thermopiles (Myceliophthora fergusii) carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD) | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, Carbohydrate esterase family 2 (CE2), GLYCEROL | | Authors: | Stogios, P.J, Dong, A, Xu, X, Cui, H, Savchenko, A. | | Deposit date: | 2015-01-27 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9449 Å) | | Cite: | Crystal structure of a Corynascus thermopiles carbohydrate esterase family 2 (CE2) enzyme plus carbohydrate binding domain (CBD)
To Be Published
|
|
7ZYS
 
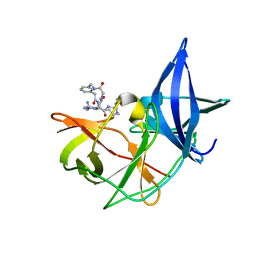 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2227 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-18-(4-azanylbutyl)-15-butyl-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(26),22,24-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-25 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
7W0Z
 
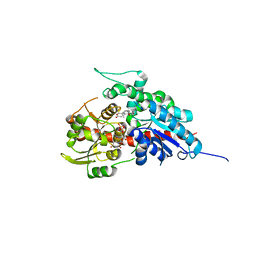 | | Glycosyltranferase UGT74AN2 | | Descriptor: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wei, H, Feng, L. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and Structural Dissection of a Plant Steroid 3-O-Glycosyltransferase Facilitated the Engineering Enhancement of Sugar Donor Promiscuity
Acs Catalysis, 2022
|
|
7ZYV
 
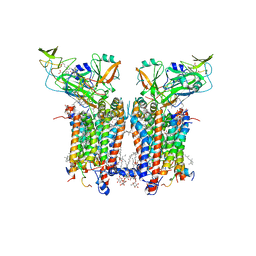 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
3L30
 
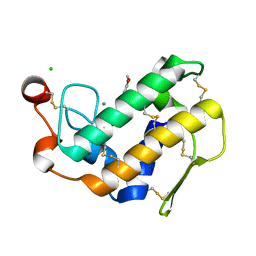 | | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine | | Descriptor: | 4,14-dihydro-8H-[1,3]dioxolo[4,5-g]isoquino[3,2-a]isoquinoline-9,10-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Naveen, C, Prasanth, G.K, Abhilash, J, Pradeep, M, Ponnuraj, K, Sadasivan, C, Haridas, M. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine
To be Published
|
|
3GPE
 
 | | Crystal Structure Analysis of PKC (alpha)-C2 domain complexed with Ca2+ and PtdIns(4,5)P2 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Protein kinase C alpha type, ... | | Authors: | Ferrer-Orta, C, Querol-Audi, J, Fita, I, Verdaguer, N. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8A15
 
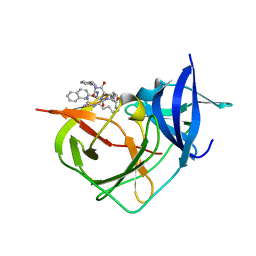 | | Crystal Structure of Unlinked NS2B-NS3 Protease from Zika Virus in Complex with Inhibitor MI-2230 | | Descriptor: | 1-[(8~{R},15~{S},18~{S})-15-(4-azanylbutyl)-18-(naphthalen-2-ylmethyl)-4,7,14,17,20-pentakis(oxidanylidene)-3,6,13,16,19-pentazabicyclo[20.3.1]hexacosa-1(25),22(26),23-trien-8-yl]guanidine, Serine protease NS3, Serine protease subunit NS2B | | Authors: | Huber, S, Steinmetzer, T. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Synthesis and structural characterization of new macrocyclic inhibitors of the Zika virus NS2B-NS3 protease.
Arch Pharm, 2024
|
|
3G60
 
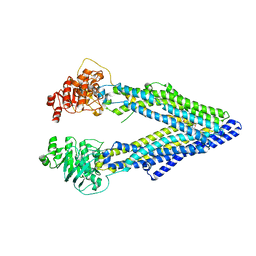 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
6PUU
 
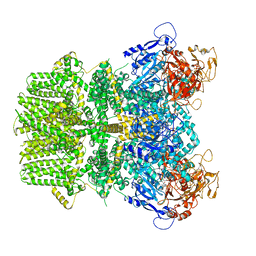 | | Human TRPM2 bound to 8-Br-cADPR and calcium | | Descriptor: | (2R,3R,4S,5R,13R,14S,15R,16R)-24-amino-18-bromo-3,4,14,15-tetrahydroxy-7,9,11,25,26-pentaoxa-17,19,22-triaza-1-azonia-8 ,10-diphosphapentacyclo[18.3.1.1^2,5^.1^13,16^.0^17,21^]hexacosa-1(24),18,20,22-tetraene-8,10-diolate 8,10-dioxide, CALCIUM ION, Transient receptor potential cation channel subfamily M member 2 | | Authors: | Du, J, Lu, W, Huang, Y. | | Deposit date: | 2019-07-18 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ligand recognition and gating mechanism through three ligand-binding sites of human TRPM2 channel.
Elife, 8, 2019
|
|
3G61
 
 | | Structure of P-glycoprotein Reveals a Molecular Basis for Poly-Specific Drug Binding | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1a | | Authors: | Aller, S.G, Yu, J, Ward, A, Weng, Y, Chittaboina, S, Zhuo, R, Harrell, P.M, Trinh, Y.T, Zhang, Q, Urbatsch, I.L, Chang, G. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding.
Science, 323, 2009
|
|
6GXC
 
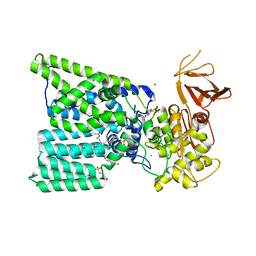 | | Bacterial oligosaccharyltransferase PglB in complex with an inhibitory peptide and a reactive lipid-linked oligosaccharide analog | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLY-ASP-GLN-DAB-ALA-THR-PPN-GLY, ... | | Authors: | Napiorkowska, M, Locher, K.P, Boilevin, J, Darbre, T, Reymond, J.-L. | | Deposit date: | 2018-06-27 | | Release date: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structure of bacterial oligosaccharyltransferase PglB bound to a reactive LLO and an inhibitory peptide.
Sci Rep, 8, 2018
|
|
6GYR
 
 | | Transcription factor dimerization activates the p300 acetyltransferase | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Panne, D, Ortega, E. | | Deposit date: | 2018-07-01 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Transcription factor dimerization activates the p300 acetyltransferase.
Nature, 562, 2018
|
|
7X81
 
 | | The crystal structure of PloI4-C16M/D46A/I137V in complex with exo-2+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,11E,12aR,14aS,17E,18aR,18bR)-9-ethyl-4,8,19-trihydroxy-11,12a,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,12a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)cyclobuta[b]naphtho[2,1-j][1]azacyclotetradecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-10 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
7X86
 
 | | The crystal structure of PloI4-F124L in complex with endo-4+2 adduct | | Descriptor: | (4S,4aS,6aR,8R,9R,10aS,13S,14aS,18aR,18bR,E)-9-ethyl-4,8,19-trihydroxy-10a,12,13,18a-tetramethyl-2,3,4,4a,6a,7,8,9,10,10a,13,14,18a,18b-tetradecahydro-14a,17-(metheno)benzo[b]naphtho[2,1-h][1]azacyclododecine-16,18(1H,15H)-dione, PloI4 | | Authors: | Li, M, Pan, L.F. | | Deposit date: | 2022-03-11 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | A cyclase that catalyses competing 2 + 2 and 4 + 2 cycloadditions.
Nat.Chem., 15, 2023
|
|
3MP3
 
 | | Crystal Structure of Human Lyase in complex with inhibitor HG-CoA | | Descriptor: | (3R,5S,9R,21S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9,21-tetrahydroxy-8,8-dimethyl-10,14,19-trioxo-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatricosan-23-oic acid 3,5-dioxide, 3-HYDROXYPENTANEDIOIC ACID, Hydroxymethylglutaryl-CoA lyase, ... | | Authors: | Fu, Z, Runquist, J.A, Montgomery, C, Miziorko, H.M, Kim, J.-J.P. | | Deposit date: | 2010-04-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights into human HMG-CoA lyase from structures of Acyl-CoA-containing ternary complexes.
J.Biol.Chem., 285, 2010
|
|
3M5L
 
 | | Crystal structure of HCV NS3/4A protease in complex with ITMN-191 | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3/4A, SULFATE ION, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3FSN
 
 | | Crystal structure of RPE65 at 2.14 angstrom resolution | | Descriptor: | FE (II) ION, Retinal pigment epithelium-specific 65 kDa protein, TETRAETHYLENE GLYCOL | | Authors: | Kiser, P.D, Lodowski, D.T, Palczewski, K. | | Deposit date: | 2009-01-11 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of native RPE65, the retinoid isomerase of the visual cycle.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
