8T62
 
 | |
8T61
 
 | |
8T60
 
 | |
8T5Z
 
 | |
8T5Y
 
 | |
8T5X
 
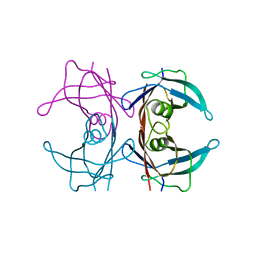 | | Probing the dissociation pathway of a kinetically labile transthyretin mutant (A25T) | | Descriptor: | Transthyretin | | Authors: | Ferguson, J.A, Sun, X, Leach, B.I, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Probing the Dissociation Pathway of a Kinetically Labile Transthyretin Mutant.
J.Am.Chem.Soc., 146, 2024
|
|
8T5W
 
 | |
8T5U
 
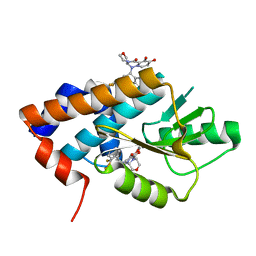
 | | ATP-1 state of Bcs1 (C7 symmetrized) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Mitochondrial chaperone BCS1 | | Authors: | Zhan, J, Xia, D. | | Deposit date: | 2023-06-14 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Conformations of Bcs1L undergoing ATP hydrolysis suggest a concerted translocation mechanism for folded iron-sulfur protein substrate.
Nat Commun, 15, 2024
|
|
8T5T
 
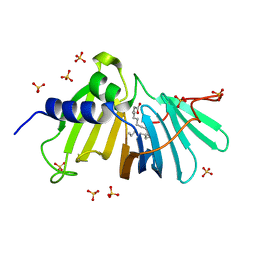 | |
8T5S
 
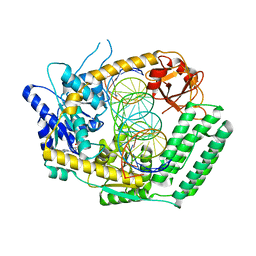 | | Cryo-EM structure of DRH-1 helicase and C-terminal domain bound to dsRNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-related helicase, MAGNESIUM ION, ... | | Authors: | Consalvo, C.D, Donelick, H.M, Shen, P.S, Bass, B.L. | | Deposit date: | 2023-06-14 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA.
Elife, 13, 2024
|
|
8T5R
 
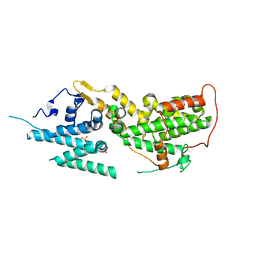 | | SOS2 crystal structure with fragment bound (compound 13) | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5Q
 
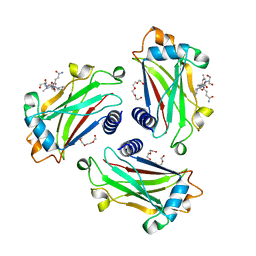 | |
8T5P
 
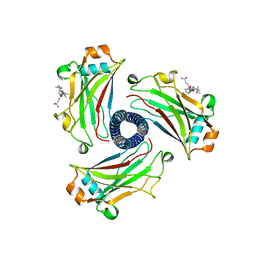 | |
8T5N
 
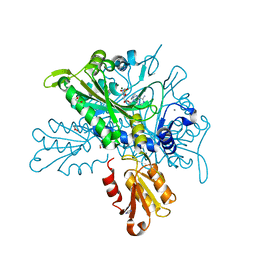 | |
8T5M
 
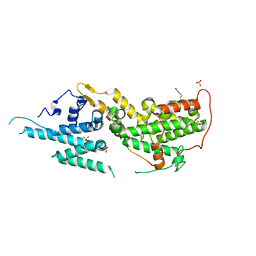 | | SOS2 crystal structure with fragment bound (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5L
 
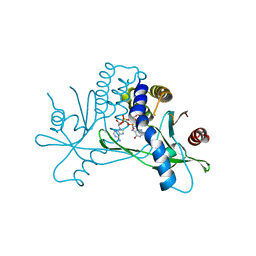 | | Crystal structure of STING CTD in complex with 2'3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5K
 
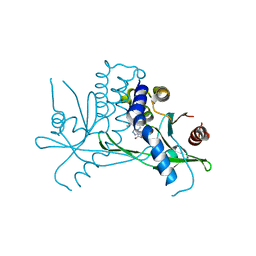 | | Crystal structure of STING CTD in complex with BDW-OH | | Descriptor: | Stimulator of interferon genes protein, {[(4S)-8,9-dimethylthieno[3,2-e][1,2,4]triazolo[4,3-c]pyrimidin-3-yl]sulfanyl}acetic acid | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
8T5J
 
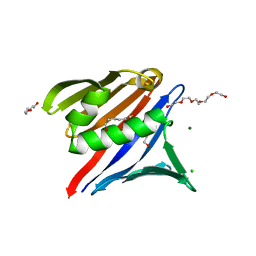 | |
8T5I
 
 | | Crystal structure of human WDR5 in complex with MR4397 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, N-[(2S)-1-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-1-oxopentan-2-yl]-3-[(1H-imidazol-1-yl)methyl]benzamide, ... | | Authors: | Kimani, S, Dong, A, Li, F, Loppnau, P, Ackloo, S, Vedadi, M, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human WDR5 in complex with MR4397
To be published
|
|
8T5G
 
 | | SOS2 co-crystal structure with fragment bound (compound 12) | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Son of sevenless homolog 2, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5F
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Parathyroid hormone | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8T5E
 
 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | Bcl-2-like protein 11, Bim_fulldiff | | Authors: | Torres, S.V, Leung, P.J.Y, Bera, A.K, Baker, D, Kang, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
8T5C
 
 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
8T5B
 
 | |
8T5A
 
 | |
