1PJ3
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
4IHB
 
 | |
3ACD
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with IMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Hypoxanthine-guanine phosphoribosyltransferase, INOSINIC ACID | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1OCW
 
 | |
2ZTH
 
 | |
2ZLB
 
 | |
1RQK
 
 | | Structure of the reaction centre from Rhodobacter sphaeroides carotenoidless strain R-26.1 reconstituted with 3,4-dihydrospheroidene | | Descriptor: | 3,4-DIHYDROSPHEROIDENE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Roszak, A.W, Hashimoto, H, Gardiner, A.T, Cogdell, R.J, Isaacs, N.W. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein Regulation of Carotenoid Binding: Gatekeeper and Locking Amino Acid Residues in Reaction Centers of Rhodobacter sphaeroides
STRUCTURE, 12, 2004
|
|
3A6J
 
 | | E122Q mutant creatininase complexed with creatine | | Descriptor: | Creatinine amidohydrolase, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
2GH7
 
 | | Epi-biotin complex with core streptavidin | | Descriptor: | BIOTIN, EPI-BIOTIN, GLYCEROL, ... | | Authors: | Le Trong, I, Aubert, D.G.L, Thomas, N.R, Stenkamp, R.E. | | Deposit date: | 2006-03-26 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The high-resolution structure of (+)-epi-biotin bound to streptavidin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1ONN
 
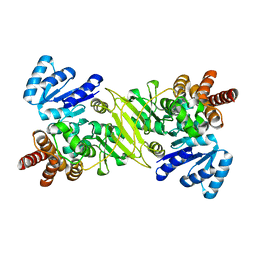 | | IspC apo structure | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
2RSY
 
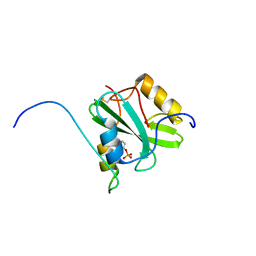 | | Solution structure of the SH2 domain of Csk in complex with a phosphopeptide from Cbp | | Descriptor: | Phosphoprotein associated with glycosphingolipid-enriched microdomains 1, Tyrosine-protein kinase CSK | | Authors: | Tanaka, H, Akagi, K, Oneyama, C, Tanaka, M, Sasaki, Y, Kanou, T, Lee, Y, Yokogawa, D, Debenecker, M, Nakagawa, A, Okada, M, Ikegami, T. | | Deposit date: | 2012-09-10 | | Release date: | 2013-04-10 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Identification of a new interaction mode between the Src homology 2 domain of C-terminal Src kinase (Csk) and Csk-binding protein/phosphoprotein associated with glycosphingolipid microdomains.
J.Biol.Chem., 288, 2013
|
|
1XGD
 
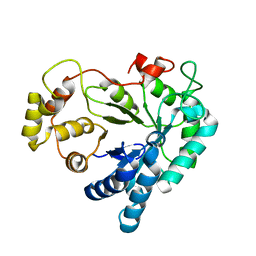 | | Apo R268A human aldose reductase | | Descriptor: | Aldose reductase | | Authors: | Brownlee, J.M, Bohren, K.M, Milne, A.C, Gabbay, K.H, Harrison, D.H.T. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Apo R268A human aldose reductase: Hinges and latches that control the kinetic mechanism
Biochim.Biophys.Acta, 1748, 2005
|
|
4H1N
 
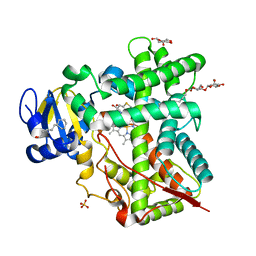 | | Crystal Structure of P450 2B4 F297A Mutant in Complex with Anti-platelet Drug Clopidogrel | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Clopidogrel, Cytochrome P450 2B4, ... | | Authors: | Shah, M.B, Jang, H.H, Stout, C.D, Halpert, J.R. | | Deposit date: | 2012-09-10 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray crystal structure of the cytochrome P450 2B4 active site mutant F297A in complex with clopidogrel: Insights into compensatory rearrangements of the binding pocket.
Arch.Biochem.Biophys., 530, 2013
|
|
1V1B
 
 | |
1PJ4
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate malate, ATP, Mn++, and allosteric activator fumarate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, D-MALATE, FUMARIC ACID, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-31 | | Release date: | 2003-09-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1ONP
 
 | | IspC complex with Mn2+ and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
5LGR
 
 | | Crystal structure of mouse CARM1 in complex with ligand P1C3u | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Marechal, N, Troffer-Charlier, N, Cura, V, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state mimics are valuable mechanistic probes for structural studies with the arginine methyltransferase CARM1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LIG
 
 | | G-Quadruplex formed at the 5'-end of NHEIII_1 Element in human c-MYC promoter bound to triangulenium based fluorescence probe DAOTA-M2 | | Descriptor: | 8,12-bis(2-morpholinoethyl)-8H-benzo[ij]xantheno[1,9,8-cdef][2,7]naphthyridin-12-iumhexafluorophosphate, DNA (5'-D(*TP*AP*GP*GP*GP*AP*GP*GP*GP*TP*AP*GP*GP*GP*AP*GP*GP*GP*T)-3') | | Authors: | Kotar, A, Wang, B, Shivalingam, A, Gonzalez-Garcia, J, Vilar, R, Plavec, J. | | Deposit date: | 2016-07-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Triangulenium-Based Long-Lived Fluorescence Probe Bound to a G-Quadruplex.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
4LQW
 
 | |
1BFF
 
 | | THE 154 AMINO ACID FORM OF HUMAN BASIC FIBROBLAST GROWTH FACTOR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR, BETA-MERCAPTOETHANOL, PHOSPHATE ION | | Authors: | Kastrup, J.S, Eriksson, E.S. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of the 154-amino-acid form of recombinant human basic fibroblast growth factor. comparison with the truncated 146-amino-acid form.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
7LTO
 
 | | Nse5-6 complex | | Descriptor: | Non-structural maintenance of chromosome element 5, Ubiquitin-like protein SMT3,DNA repair protein KRE29 chimera | | Authors: | Yu, Y, Li, S.B, Zheng, S, Tangy, S, Koyi, C, Wan, B.B, Kung, H.H, Andrej, S, Alex, K, Patel, D.J, Zhao, X.L. | | Deposit date: | 2021-02-19 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Integrative analysis reveals unique structural and functional features of the Smc5/6 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5M19
 
 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6WPP
 
 | | HUMAN NIK IN COMPLEX WITH LIGAND COMPOUND X | | Descriptor: | 5-fluoro-N-methyl-2-(7H-pyrrolo[2,3-d]pyrimidin-5-yl)pyrimidin-4-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Shih, A.Y, Hack, M, Mirzadegan, T. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Impact of Protein Preparation on Resulting Accuracy of FEP Calculations.
J.Chem.Inf.Model., 60, 2020
|
|
3U5O
 
 | |
