1ECX
 
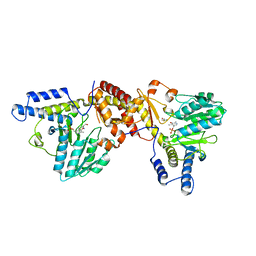 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, CYSTEINE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Kaiser, J.T, Clausen, T.C, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-01-26 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
1EG5
 
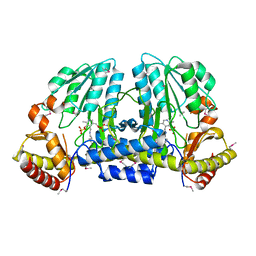 | | NIFS-LIKE PROTEIN | | Descriptor: | AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Kaiser, J.T, Clausen, T, Bourenkow, G.P, Bartunik, H.-D, Steinbacher, S, Huber, R. | | Deposit date: | 2000-02-13 | | Release date: | 2000-04-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a NifS-like protein from Thermotoga maritima: implications for iron sulphur cluster assembly.
J.Mol.Biol., 297, 2000
|
|
2AEV
 
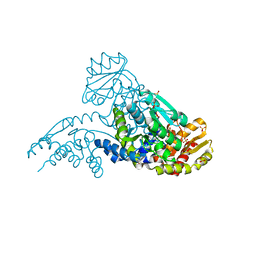 | | MJ0158, NaBH4-reduced form | | Descriptor: | Hypothetical protein MJ0158, SULFATE ION | | Authors: | Kaiser, J.T, Gromadski, K, Rother, M, Engelhardt, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-07-24 | | Release date: | 2005-10-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional investigation of a putative archaeal selenocysteine synthase
Biochemistry, 44, 2005
|
|
2AEU
 
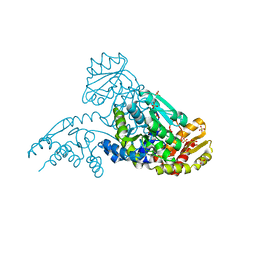 | | MJ0158, apo form | | Descriptor: | Hypothetical protein MJ0158, SULFATE ION | | Authors: | Kaiser, J.T, Gromadski, K, Rother, M, Engelhardt, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-07-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional investigation of a putative archaeal selenocysteine synthase
Biochemistry, 44, 2005
|
|
3PDI
 
 | | Precursor bound NifEN | | Descriptor: | IRON/SULFUR CLUSTER, L-Cluster (Fe8S9), Nitrogenase MoFe cofactor biosynthesis protein NifE, ... | | Authors: | Kaiser, J.T, Hu, Y, Wiig, J.A, Rees, D.C, Ribbe, M.W. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Science, 331, 2011
|
|
1N2T
 
 | | C-DES Mutant K223A with GLY Covalenty Linked to the PLP-cofactor | | Descriptor: | GLYCINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
1N31
 
 | | Structure of A Catalytically Inactive Mutant (K223A) of C-DES with a Substrate (Cystine) Linked to the Co-Factor | | Descriptor: | CYSTEINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-25 | | Release date: | 2003-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
4WZB
 
 | | Crystal Structure of MgAMPPCP-bound Av2-Av1 complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Mustafi, D, Walton, M.Y, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Nitrogenase complexes: multiple docking sites for a nucleotide switch protein.
Science, 309, 2005
|
|
4WES
 
 | | Nitrogenase molybdenum-iron protein from Clostridium pasteurianum at 1.08 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, ... | | Authors: | Zhang, L.M, Morrison, C.N, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2014-09-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 angstrom resolution: comparison with the Azotobacter vinelandii MoFe protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2O1I
 
 | | RH(BPY)2CHRYSI complexed to mismatched DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*G)-3', bis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)[chrysene-5,6-diiminato(2-)-kappa~2~N,N']rhodium(4+) | | Authors: | Pierre, V.C, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2006-11-28 | | Release date: | 2007-01-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Insights into finding a mismatch through the structure of a mispaired DNA bound by a rhodium intercalator.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4E1U
 
 | | [Ru(bpy)2 dppz]2+ bound to DNA | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*AP*TP*TP*AP*CP*CP*G)-3', BARIUM ION, Delta-[Ru(bpy)2dppz]2+ | | Authors: | Song, H, Kaiser, J.T, Barton, J.K. | | Deposit date: | 2012-03-07 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Crystal structure of delta-[Ru(bpy)2dppz]2+ bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation.
Nat Chem, 4, 2012
|
|
6MJR
 
 | | Azurin 122W/124F/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
6MJS
 
 | | Azurin 122W/124W/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
4WZA
 
 | | Asymmetric Nucleotide Binding in the Nitrogenase Complex | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, ... | | Authors: | Tezcan, F.A, Kaiser, J.T, Howard, J.B, Rees, D.C. | | Deposit date: | 2014-11-19 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8995 Å) | | Cite: | Structural evidence for asymmetrical nucleotide interactions in nitrogenase.
J.Am.Chem.Soc., 137, 2015
|
|
2VSD
 
 | | crystal structure of CHIR-AB1 | | Descriptor: | CHIR AB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Arnon, T.I, Kaiser, J.T, Bjorkman, P.J. | | Deposit date: | 2008-04-22 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of Chir-Ab1: A Primordial Avian Classical Fc Receptor.
J.Mol.Biol., 381, 2008
|
|
6MJT
 
 | | Azurin 122F/124W/126Re | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | Authors: | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | Deposit date: | 2018-09-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
6OJA
 
 | | Crystal structure of the N. meningitides methionine-binding protein in its L-methionine bound conformation | | Descriptor: | Lipoprotein, METHIONINE | | Authors: | Nguyen, P.T, Lai, J.Y, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2019-04-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers.
Protein Sci., 28, 2019
|
|
6CVL
 
 | | Crystal structure of the Escherichia coli ATPgS-bound MetNI methionine ABC transporter in complex with its MetQ binding protein | | Descriptor: | IODIDE ION, MERCURY (II) ION, MetI transmembrane subunit, ... | | Authors: | Nguyen, P.T, Kaiser, J.T, Rees, D.C. | | Deposit date: | 2018-03-28 | | Release date: | 2018-11-14 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.953 Å) | | Cite: | Noncanonical role for the binding protein in substrate uptake by the MetNI methionine ATP Binding Cassette (ABC) transporter.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4PKX
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
4PKE
 
 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | Descriptor: | PLATINUM (II) ION, Protein C10C5.1, isoform i | | Authors: | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | Deposit date: | 2014-05-14 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
1U40
 
 | | IspF with 4-diphosphocytidyl-2C-methyl-D-erythritol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1U3L
 
 | | IspF with Mg and CDP | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hechst, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1U43
 
 | | IspF with 4-diphosphocytidyl-2c-methyl-D-erythritol 2-phosphate | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-DIPHOSPHOCYTIDYL-2-C-METHYL-D-ERYTHRITOL 2-PHOSPHATE | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1U3P
 
 | | IspF native | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ZINC ION | | Authors: | Steinbacher, S, Kaiser, J, Wungsintaweekul, J, Hecht, S, Eisenreich, W, Gerhardt, S, Bacher, A, Rohdich, F. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of 2C-Methyl-D-Erythritol-2,4-Cyclodiphosphate Synthase Involved in Mevalonate Independent Biosynthesis of Isoprenoids
J.Mol.Biol., 316, 2002
|
|
1T3J
 
 | | Mitofusin domain HR2 V686M/I708M mutant | | Descriptor: | mitofusin 1 | | Authors: | Koshiba, T, Detmer, S.A, Kaiser, J.T, Chen, H, McCaffery, J.M, Chan, D.C. | | Deposit date: | 2004-04-26 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of mitochondrial tethering by mitofusin complexes
Science, 305, 2004
|
|
